

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.10.17670 | OTHER | 0.999987 | 0.000011 | 0.000002 |
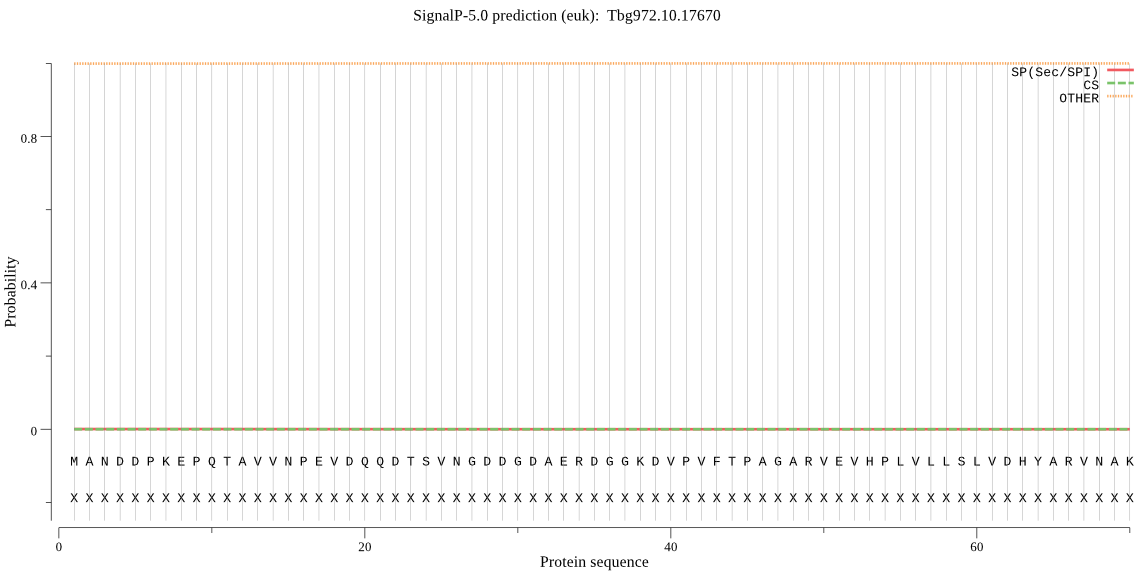
MANDDPKEPQTAVVNPEVDQQDTSVNGDDGDAERDGGKDVPVFTPAGARVEVHPLVLLSL VDHYARVNAKVSQKRRVAGLLLGRHLSHPGGIQVLDINNSFAVPFDEDPQNPDVWFLDTN YAEEMFLMFRRVHPKVRIVGWYSAGPSCAVQPNDMLLHLLIADRFSLNPVYCIVNTDPNN KGVPVLAYTTVHGRDGARSLEFRNIPTHLGAEEAEEIGIEHLLRDLTDSTITTLSAQVQE RELSLGHLGRVLQSIEEYLNDVAEGTMPISEDVLSVLQEVISLQPHIYHLKTSTDMVRHA NDQAIAMFIAAIGRCVCTLYDVIANRRRIAREIQEVRARRERSLKEKLGNEQHKAKEAAT EESRKEGAAKNKNNGGTSEGK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.10.17670 | 343 S | RRERSLKEK | 0.997 | unsp | Tbg972.10.17670 | 363 S | ATEESRKEG | 0.995 | unsp |