

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
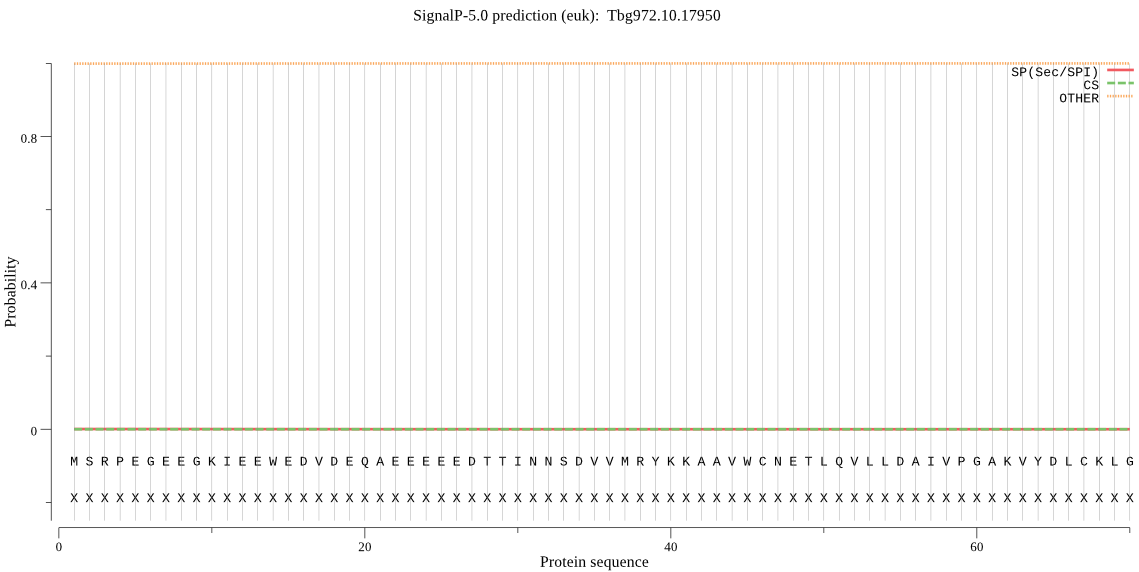
| Tbg972.10.17950 | OTHER | 0.999963 | 0.000019 | 0.000018 |
MSRPEGEEGKIEEWEDVDEQAEEEEEDTTINNSDVVMRYKKAAVWCNETLQVLLDAIVPG AKVYDLCKLGDETIAKKLRTMFRGTEKGIAFPTCISLNNCVAHNCPSPGDDAASQVVQLG DVVHVDLGIHIDGYCAQVAHTVQVTENNELGSDEKAAKVITAAYKILNTAVRKMRPGTNV YEVTEVIEKAAAHYGVTPAEGVLSHMLKRYIVDSFRCIPQKKVAEHLVHDYTLEAGQVWT LDIVMSSGKGKLKERDLRPSVFKVALDSKYTMKMESARELQREIESKYGTFPFAFRNLET KKARLGLSEMVKHSAVVPYPVLYEKDGEVVGHFKVTLLVTAKKIEVVTGLKMQKAPELPP YTDELLLTTSKLPFSLEKKRKE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| Tbg972.10.17950 | 276 S | MKMESAREL | 0.991 | unsp |