

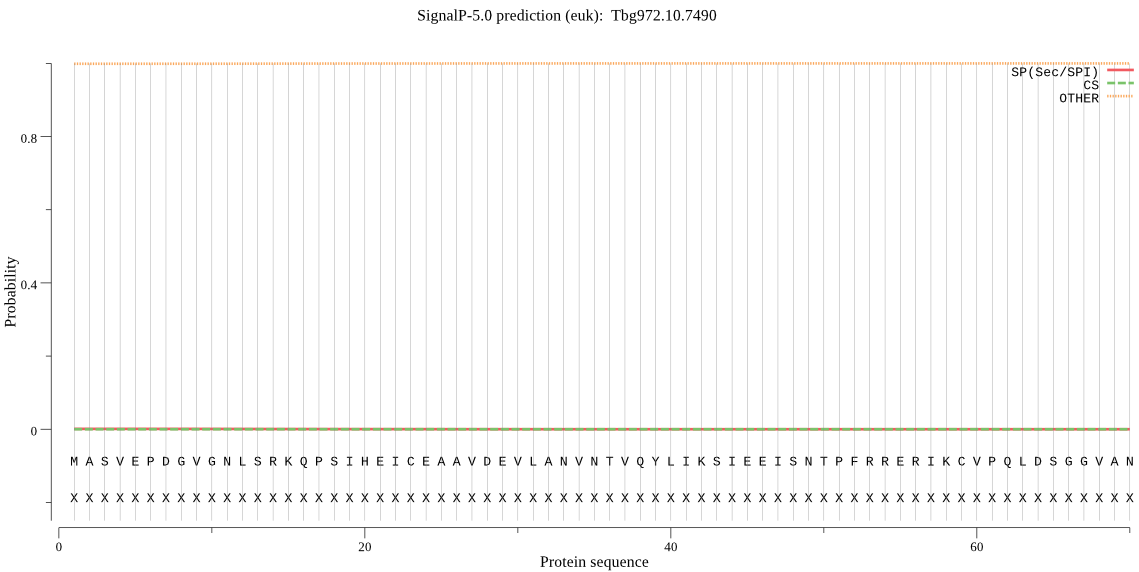
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.10.7490 | OTHER | 0.999883 | 0.000106 | 0.000011 |
MASVEPDGVGNLSRKQPSIHEICEAAVDEVLANVNTVQYLIKSIEEISNTPFRRERIKCV PQLDSGGVANASWAGGSNTMADGTSAAGYMWRRARRDCEKGDILLIEQHVVLDKATENDS IVPGAKTCTAVERNLRHELIHAFDDARGIIEASDCMHQACSEVRAARLSGDCFVGEEMRR GRFDLLSGGIQCVRRRAITAVEKNPLCRGFSERAVERIFKQCYSDYEPFAAPLYSMGSYG DEKFEL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| Tbg972.10.7490 | 18 S | RKQPSIHEI | 0.993 | unsp |