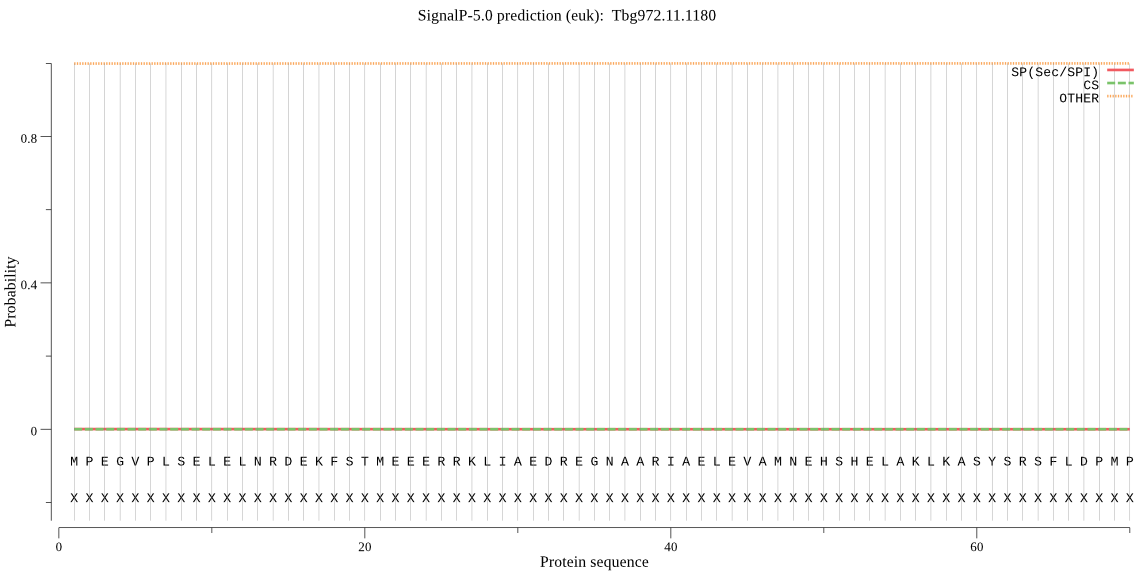
Fasta :-
MPEGVPLSELELNRDEKFSTMEEERRKLIAEDREGNAARIAELEVAMNEHSHELAKLKAS
YSRSFLDPMPEGVPLSELELDKDEKFSTMEEERRKLIAEDREGNAARIAELEVAMNEHSH
ELAKLKASYSRSFLDPMPEGVPLSELELDKDEKFSTMEEERRKLIAEDREGNATRIGRLE
RKMGRRVDKLAKLKASYSRSFLDPMPEGVPLSELELDKDEKFSTMEEERRKLIAEDREGN
AARIAELEVAMNEHSHELAKLKASYSRSFLDPMPEGVPLSELELDKDEKFSTMEEERRKL
IAEDREGNATRIAELEECMSALVRNLVICKSTLGRVIVDERIGLVPLYLLPLGEDSVLSS
LTSKLESMRCGGCLPDSVAVRDIQEQIICRVEKLAEVYLEHELDFLETVDGISVSQIALS
DDKVFCEMQRSLRQLRKSPVRNAEAIRDIEYAMNNIVRELSSKLLLEDCSYLNQCPHGVP
LADLPLDNRLFREIELKRRRLKEDDSLKNATRIRELEMKLNDHAERLAALMLSEERAFLE
PCSHGLPLSRLQLGEDSSFLAMERERRKLLREGGKDKNRIRYLEECMREHVSNVALSLIS
WKDVQFHEANHSVAEMWPRIGELYPEGIRNSVFPEITQPGDTSSAPGELGYLAPFIAALS
QHPPLIHRLLETKTHPVNGPYSFIFFDPNSNPVRVDIDDRVPVDAKCEPKFTRVPHRSWY
PLLLEKAYAKFVGGYAKLDQCTPHETLHDLTGRPVTHIPFDEKRADGIKIGDFRSVKFWK
GIKRDLAAGDVIMAMTNGSVPDGLHSQCSYALLSVIETVHESNDVSDIVIKLHNCYYDSP
TYDGPLCNDDDNWTDGLKRLCHYNPEEDALYIPVPVFLRNFSSMQRCHINCGDRLTAPGE
WTGVSCGGNPKFTTFRNNPIYLVENKTSRPVTILAELRHQAPVFFDADGVNHYHQSGLAL
LQQHHVSEIITWLLTSTTHRFLQKGILMDTREICSRMEIPPNTTCYLVPYTLKRGSYGKF
NVSLYPASSDITLVPLAALCNSHVVINVDVLLKPGSREGRRVDFSVHEACDVHLLLHQNK
ITDPASVKRGNLLAEDEVSMSVNDERMSVLATTRDASNAREQSLALELPTAGRYSVFIEC
RSRLRTGECPCTLSIYSPSRAKTRLAPLPPSGTQSVLPAIAPRQPPRQPALYGRRLDQPP
LSAGTVRAAAATLPVGLVPAPPKSGTTPQRR