

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
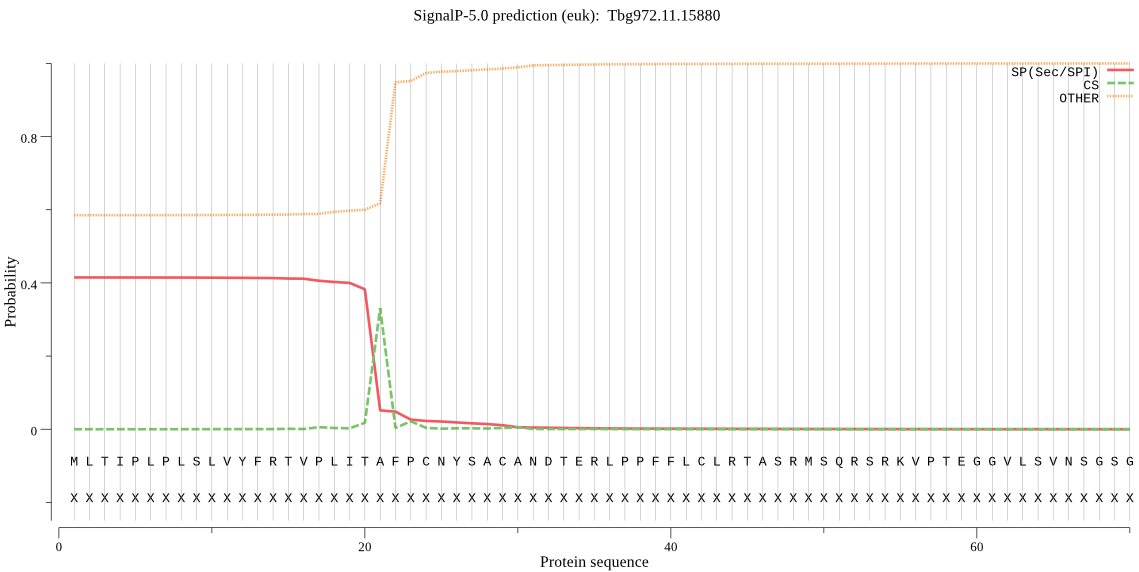
| Tbg972.11.15880 | SP | 0.166897 | 0.782424 | 0.050679 | CS pos: 21-22. ITA-FP. Pr: 0.9409 |
MLTIPLPLSLVYFRTVPLITAFPCNYSACANDTERLPPFFLCLRTASRMSQRSRKVPTEG GVLSVNSGSGLAIWNELQIQQENWGGNLGLPAREPGAVVELASTEGKIVVSTDPEQRNIF KECILGLSGSVGDITCTDFEVNWKRPVEIFAPFRPLVHRNVTPFFDPYDTQRVIGSRVPA MTSPSRRADSKSSAAAIPLHIQFPEIVTTAEFPTPEAPATESLAFSEEGVVGQYRKVQPL TQAYDVGLELGCAATPAEACGSRYSTGLRRRLNQADAISRLSEPPPFVMDAFNTAIRFVE SEQNILTKGNYLWELIHPHAPGTCHPVYNPYGKYVVRLFIDGYFRQVPIDDYLPVDALGR PFFSVTAQKEIWPALIAKALFVALGPNRHLLFTDSEAIIACLLGEWVPQRINPRTQPATA CALLLNARKDQLYNDSDATHFSASQVLTPDEDKGVSVSTQEQCNTSPWVDNQINERPASQ SKRASRLEASFANGANSPLSYDISVTNVPAVPETPVICAVGVLPHDMRKLYVIHKVVVFR DTLALKISTNPPSSIIEPKELTLDNKDEAVQGLLHHPSCGDLDQRGTGWAASQESAVASS AFWVTFEELAESLDIIIWRQLGQESPFTFNSRITYTDTSSHPRKKGAASAAAAATTVARQ TVVRWMHISSKAPEQIAIVNLGGSASCRAASAVIGSPTPSLLAAAHQTPALSMMGSESSQ LKALSGEGREVHLDLYRWDRGDVLCRAASFACEPSRLECMLHTVPAGSHVFRVTVVGLEP QEVVGLFSTRNFILGDEKEALRSANIFKMSDAGAHPGVEKPNEEVIWFKHRFAVKEPTIV SFVLSTLESGEELSMYRDMPGASGKESKGVKTRNTNVRLHSKEAAEGSPPRGAHAEIIPY CNVLLVDLDAGTSKRGAVGHLVRQRIEPNQRGYLVMAYALVEADSISFSLKPADLCADSP LEYGSPREKETNGLSPCGDKGPLTPFGVRPHLFSKGWWRLVVLSSVPLDSYRAIPKDTWS FAEKGQLKQGSNALLFSYACTVSDRTDITLLLDVHSQRPIPFHIKIFRAGVEGPPVFVSE ECVNHLFVPHVAVDVPEKVKNVAYIIEAWLDKEKVEEWERMRRLSQELKFLLAAEAARVK AEMKQQKELQEYHEDPRAFKERLLNQEMGPVENLVPTPLREQMSVSSKKGRPSVDKRRTL SNAAPSRQTIVRTSVTPSTPASAVAVHIDSIDPELVVTYDLRLYFSAKTDVKCVATGKDP LTTMRGLWVPPIDPLFGEAPPLSGGRSGGQRGKQKDSSPSPEELYKADQGRLSRQNFLEN PRNLLFPYLPQGEKIRGVEESAPNALASTPSIALVVGVEDEPNFLHAPILDESAHRVQHF SLYATEVVPPTHGSPTQSASGVPFLRTRSPNGRIAAPTFGNNPPPTDDDALAIELKTPVN EVCLWLGEELRRQEERRAECRNGIKTLMREYWETRKPGAAVLALPHAREDEGNKRPRPRS KMSS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.11.15880 | 190 S | RRADSKSSA | 0.996 | unsp | Tbg972.11.15880 | 190 S | RRADSKSSA | 0.996 | unsp | Tbg972.11.15880 | 190 S | RRADSKSSA | 0.996 | unsp | Tbg972.11.15880 | 485 S | SKRASRLEA | 0.996 | unsp | Tbg972.11.15880 | 554 S | NPPSSIIEP | 0.997 | unsp | Tbg972.11.15880 | 863 S | MPGASGKES | 0.997 | unsp | Tbg972.11.15880 | 881 S | VRLHSKEAA | 0.996 | unsp | Tbg972.11.15880 | 965 S | LEYGSPREK | 0.996 | unsp | Tbg972.11.15880 | 1125 S | MRRLSQELK | 0.997 | unsp | Tbg972.11.15880 | 1193 S | KGRPSVDKR | 0.994 | unsp | Tbg972.11.15880 | 1209 T | PSRQTIVRT | 0.991 | unsp | Tbg972.11.15880 | 1214 S | IVRTSVTPS | 0.996 | unsp | Tbg972.11.15880 | 1298 S | QKDSSPSPE | 0.997 | unsp | Tbg972.11.15880 | 1300 S | DSSPSPEEL | 0.998 | unsp | Tbg972.11.15880 | 1500 S | PRPRSKMSS | 0.993 | unsp | Tbg972.11.15880 | 50 S | ASRMSQRSR | 0.997 | unsp | Tbg972.11.15880 | 53 S | MSQRSRKVP | 0.993 | unsp |