

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
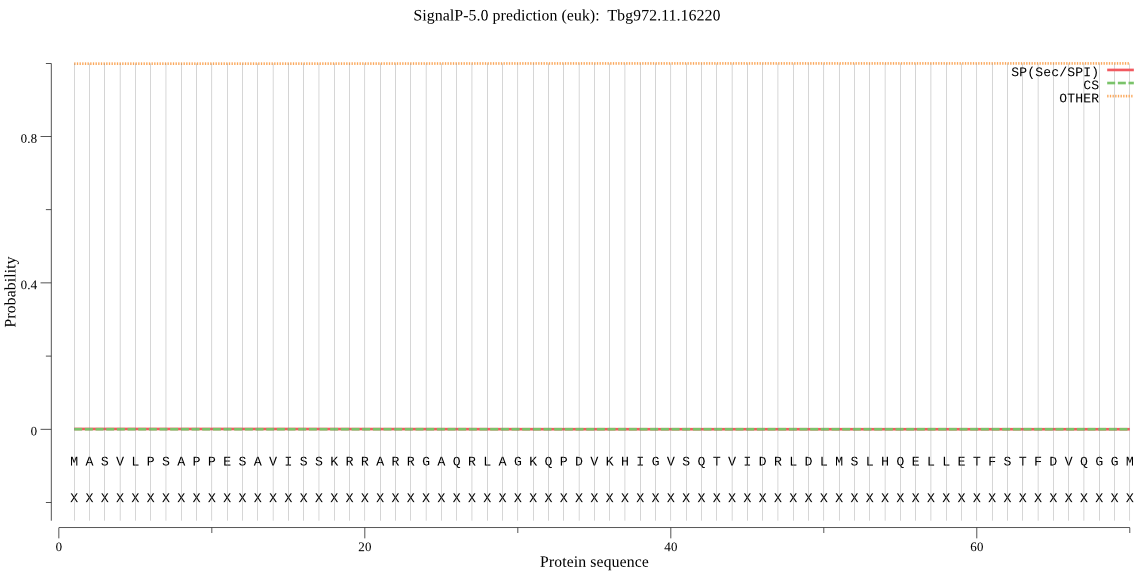
| Tbg972.11.16220 | OTHER | 0.999589 | 0.000011 | 0.000399 |
MASVLPSAPPESAVISSKRRARRGAQRLAGKQPDVKHIGVSQTVIDRLDLMSLHQELLET FSTFDVQGGMRRKGQRGSFASKFNHVPIAKGIVNRTNSCFMNAMLQAIIFTPPLAQLIIS ASDSELCPTLSALGKWMLSYWTKPANQSISPPQLSVAQSANEGVASTYLLLARMNGYAQE DALEFLQHLLDTINTELCSLEKHYCNRAAIDSGDEKGWTFVHGRERRSLREEKTGPHSIL LDAVFGGTIQSHLKGKSRVRSHASVVLDRFFVLQVDVGFNAECTLEEALERTLQTEKVYD DSRAKDLVKTMKLHQLPLVLFAQLRRWVVTAEGELVKLDNIVRFGKTLVLPKSICSDETL SGTARTYQLVAFVAHRGSATGCGHYVTYIVNASLPENARSNSLVTGGEEVLTLCNDVRIS HVTMREAIEGEAAYLLVYQRKT
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| Tbg972.11.16220 | 228 S | RERRSLREE | 0.998 | unsp |