

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.11.4220 | OTHER | 0.843854 | 0.001059 | 0.155087 |
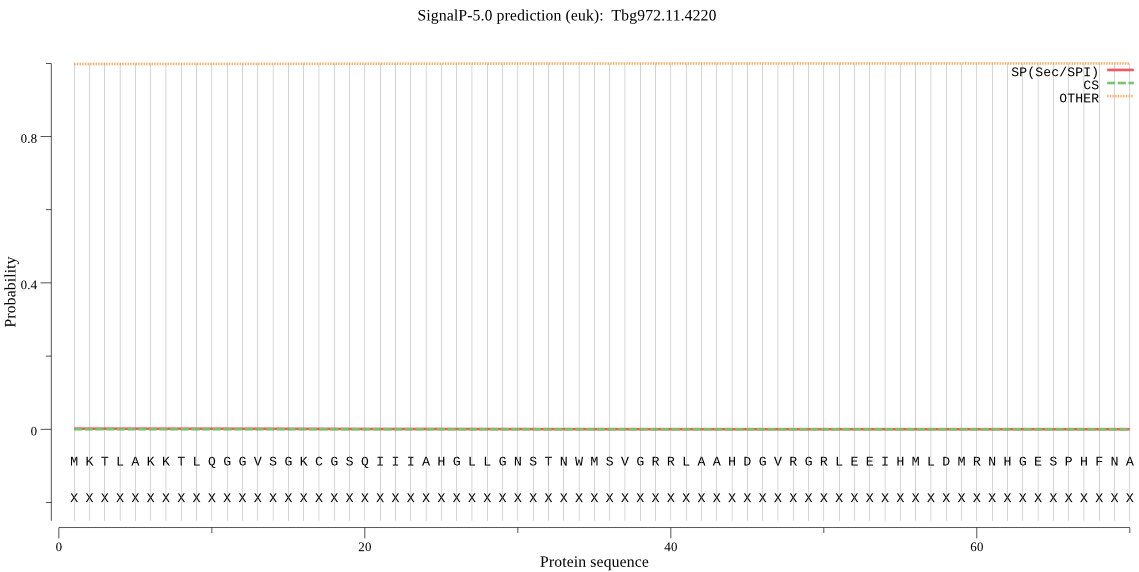
MKTLAKKTLQGGVSGKCGSQIIIAHGLLGNSTNWMSVGRRLAAHDGVRGRLEEIHMLDMR NHGESPHFNAHTNATMASDIEHFVLQQQRQWQSRAGAEGDGGIVLIGHSMGGFAVMGSML RRANETSLLLQSGVEELEQRCKRGDYYGWCDEQGNAAEMRAVNRAMGLPETQPLYDILYD CKGDNVARPPRVKAVVIVDITPSTALGTHRQHGQSSSETLDAMVAADLSRVHSFGDGNAE LERVGVSNKAMRDFLLTNLRLDPRTKEATWRCNLPVLRADYNSIALGVSGWFLSASEKVS RDRGGELVAPLRCSLPTLFVFGQNSPYNTPEDRRLIPRFFSNAIEVEVEGAGHFVHYEKM QEFVNVVVPFLTEYL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.11.4220 | 215 S | QHGQSSSET | 0.995 | unsp | Tbg972.11.4220 | 233 S | SRVHSFGDG | 0.995 | unsp |