

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.11.4340 | SP | 0.120993 | 0.878474 | 0.000532 | CS pos: 30-31. CKA-HA. Pr: 0.4561 |
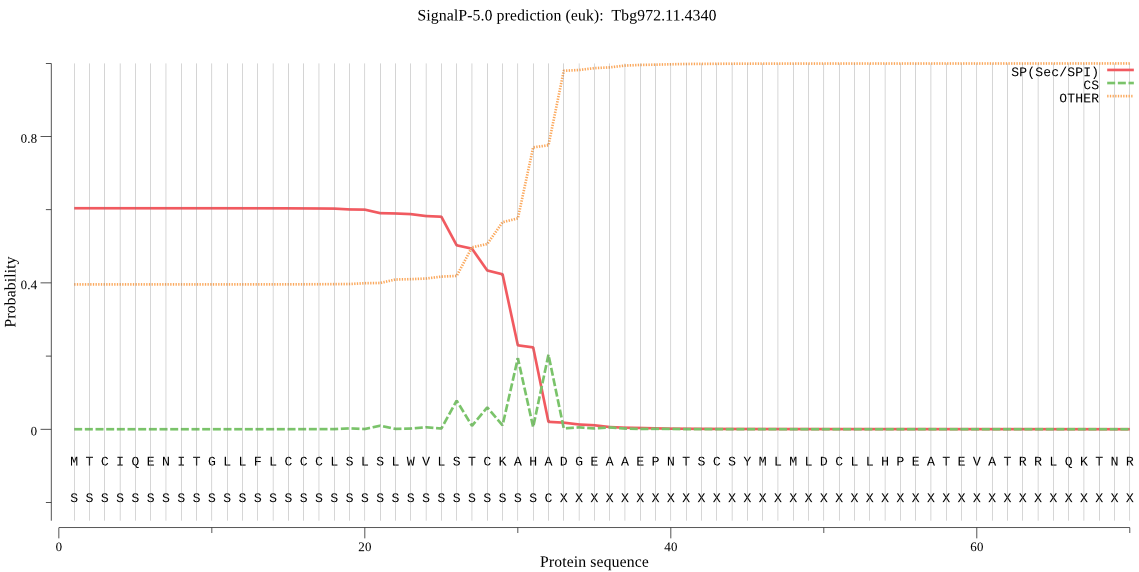
MTCIQENITGLLFLCCCLSLSLWVLSTCKAHADGEAAEPNTSCSYMLMLDCLLHPEATEV ATRRLQKTNRWKWWRSGLPSGSEVHVHPPPAPFGTWNDFLLMDIRSPLCGKLSGNVMESE ISGTSFALPTVGRVIVLRVVEDRWVPKEFTRTHSIESRGDKLSRWRYPRQYLRLRTALRW SGLGVRVAVLDNGVDAALLPQRDVEGIPNSAGEKSDWSVALCRSFVPDVPCENRRDSHGT FSVSVTAGNISLTSNLRSNEGFGNQTTSFTDASTAEEMPAVSKHYVGVAPGSTVGMFRVF DDARGSKTSWLVSALNDVLQWRADVVSLAFGGTDNMDTIFTEKIRVLAMSGVVVVAAAGN DGPTMGTIHNPADQAEVLAVGSLGTVNCRIVASVMTAHPGHDDGEGNCTDSRDQRWVSQF SGRGPSTVEFPFGAGRVRPDILALGEHVVGVGRVMEKFGMSGEERVLGLQVSHGTSVATA LVAGVAALCIEALRTLGDGTRVNVALVKGLLIETAVELVPNTESLNSVERVLMQERKKYS PQGAGSWSTEGDAFLNGKRRRTLDGVDMVHTLFHYRNVLQFSRLSQGGGEVCPTCALSRI ELRAHEYDRLLEVFAFPKAVDATADHHLPHGGKGPAAATLGPCLLNWPYCEQPLFPSAAP IAFNLSLHNARCESSRLNLISPNVTISGAEGFCSTNRVCEPGYLGAEIAQQLVLVRADAS LVMSAHSGWLSLFALSPANASTTQIPPPTSAAESNSPDSFALDRYDLITVIGSVRLAYLC TASGEHQKANADVDESGGVRSVTVPFKLPVVRRPSRLERVGFDMSHQWFYPPGQVPDDDI RQERHVLHYRRGNGRLRGHAVRSRSGEKRCGAYECESDHLYTNMAPFFLYLRRVLGLFVE QPLLTYLPFGVSKIIGFGEVNAKRDVKNVSAAALKRYYGNIGALILFDVELPLLQMERTL ISDAVRYEKLHLLVVSEWFSRSIARGTSSHSSTPGGSLFPSLKGNQNVNSSFCDADEPKV KSNGCLPKEPMVLDGSSHVPSLNALLRDVSHGKLQLDTEHVVSGELVLATCPHHSYKMPS SFWSISSWISWLWFGGAQRADGCVDRHLGEIQSAGVVLWPPTGERNKQGDESHSELGGER SSADSKPTAVCSVAKNWARGLYFLRKLKDSGGNLNCDDATNGCGDGTKQREVDVMHPVGQ SAFVPIFGFVGSGGDCLGEDGLFDNEGCMKGAPAAESLRHAGGRVVLFTDSNSFSDLPRS AATLHRLNLLIYDTSRLLSRSSTETSFAVDISHKIEAIVREESFQPSLSLGLIGDFVSFL HNGGTKNFMQGVDCRRSDGTSLGHDWNDISTSEKAENGGRDGLHAANEEDKASLLALRLF QGAPQRVAIAERVRAVLVGWEAAIASEADYTETDAVITDGDCNGSFFVQTADSRKLWSNC TYGGEGVNMILWTHILGLTFTTLVLYATNCLRCLLENMKVVAVEPSE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.11.4340 | 237 S | NRRDSHGTF | 0.997 | unsp | Tbg972.11.4340 | 237 S | NRRDSHGTF | 0.997 | unsp | Tbg972.11.4340 | 237 S | NRRDSHGTF | 0.997 | unsp | Tbg972.11.4340 | 426 S | GRGPSTVEF | 0.993 | unsp | Tbg972.11.4340 | 461 S | KFGMSGEER | 0.994 | unsp | Tbg972.11.4340 | 585 S | FSRLSQGGG | 0.99 | unsp | Tbg972.11.4340 | 815 S | VRRPSRLER | 0.996 | unsp | Tbg972.11.4340 | 865 S | VRSRSGEKR | 0.994 | unsp | Tbg972.11.4340 | 1011 S | NVNSSFCDA | 0.991 | unsp | Tbg972.11.4340 | 1132 S | QGDESHSEL | 0.995 | unsp | Tbg972.11.4340 | 1253 S | TDSNSFSDL | 0.993 | unsp | Tbg972.11.4340 | 1279 S | SRLLSRSST | 0.997 | unsp | Tbg972.11.4340 | 1281 S | LLSRSSTET | 0.994 | unsp | Tbg972.11.4340 | 1282 S | LSRSSTETS | 0.995 | unsp | Tbg972.11.4340 | 154 S | TRTHSIESR | 0.992 | unsp | Tbg972.11.4340 | 157 S | HSIESRGDK | 0.993 | unsp |