-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
-
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs: GO:0005739
-
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
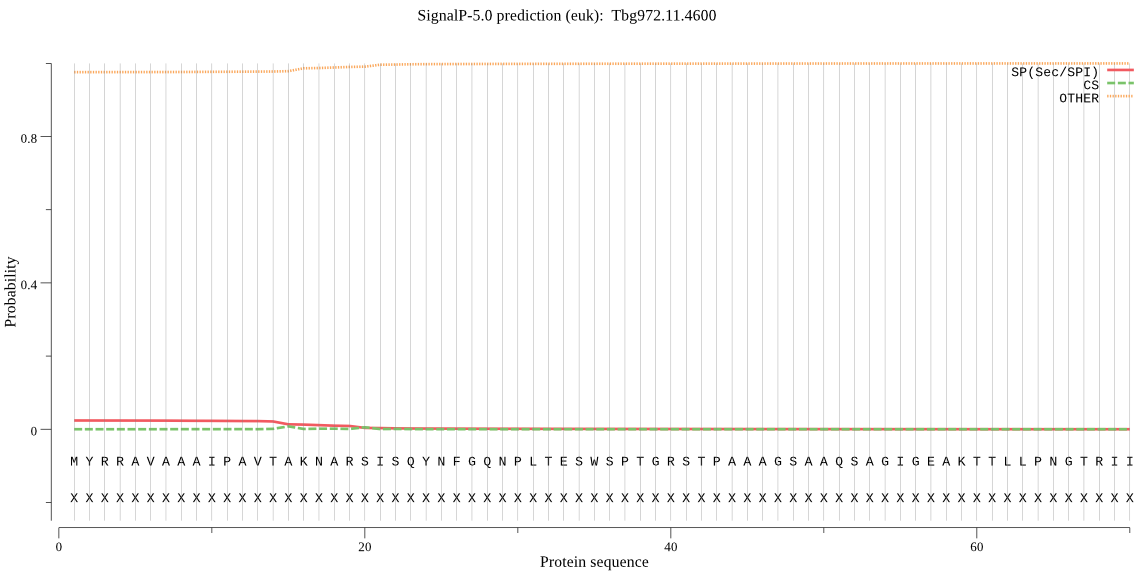
|
_ID | Prediction | OTHER | SP | mTP | CS_Position |
|
Tbg972.11.4600 | OTHER | 0.538224 | 0.071658 | 0.390118 | |
No Results
-
Fasta :-
>Tbg972.11.4600
MYRRAVAAAIPAVTAKNARSISQYNFGQNPLTESWSPTGRSTPAAAGSAAQSAGIGEAKT
TLLPNGTRIITQQRGGPVVSIGAYILAGPAYDPVGCPGLHHLMHTALTTSNYNNSLFQLD
RSIRSTGASFSHFEKDKYYIGLRLDARVDMWKSNTSGATSDAKRFDLNLLQDTIFTAISA
PRFHEPDLERFRDTIDNNLKELRWQRPAQYAIQQLETVAFYKEPLGNPRHVPEWSNGRIA
SKALLDQYARYVTPDRVVVAGVNMEHDELVAEYESNPYPHSSNAPHHAAFAKGLGKPAFD
ITNEHSQYSGGELHEHEDRPKEMCTKPDMDTETAIAVGYLAFGRSKTSLQRYAATLVYQQ
LFNIVIHDSLRYEQDVVLDGVRSFYLPYHSAGLVGFTAITSPENAVPLVKATMKGIQNVK
FDNAALLEAAKHRAAVELTSQCWDSSRDICDYLGTSLSLDAKASNSTQYLNPSEVLAAVR
NVTASELKEVKECMMGSKPSLFGHGEMLAFPSLRQLGA
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/92
Sequence name : 92
Sequence length : 518
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.918
CoefTot : -3.879
ChDiff : -3
ZoneTo : 133
KR : 10
DE : 4
CleavSite : 126
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.312 1.171 0.215 0.615
MesoH : 0.006 0.329 -0.225 0.241
MuHd_075 : 40.752 24.954 10.490 8.814
MuHd_095 : 35.269 26.128 9.091 8.215
MuHd_100 : 36.460 24.069 9.175 8.284
MuHd_105 : 38.304 23.131 9.938 8.987
Hmax_075 : 13.700 10.733 2.309 4.630
Hmax_095 : 10.675 15.200 0.438 5.210
Hmax_100 : 15.200 16.800 2.602 6.430
Hmax_105 : 10.150 13.475 1.900 4.891
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.1523 0.8477
DFMC : 0.1409 0.8591
This protein is probably imported in mitochondria.
f(Ser) = 0.1053 f(Arg) = 0.0602 CMi = 0.68259
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>Tbg972.11.4600
ATGTACCGACGCGCCGTTGCTGCTGCCATACCCGCCGTCACTGCCAAGAATGCACGGTCA
ATCTCGCAGTACAACTTTGGTCAGAACCCCCTGACTGAGTCGTGGTCACCGACTGGCCGC
TCAACTCCCGCGGCAGCCGGTTCTGCTGCGCAGTCAGCTGGCATTGGTGAGGCGAAGACG
ACGCTACTCCCTAATGGAACCCGAATCATCACGCAGCAGCGAGGTGGTCCTGTGGTGTCA
ATCGGCGCCTATATTCTTGCGGGTCCTGCTTACGATCCAGTTGGCTGTCCGGGCCTTCAT
CATCTGATGCATACCGCACTCACGACAAGCAACTACAATAACTCACTCTTCCAACTGGAC
CGCAGTATTCGCAGTACTGGTGCTTCGTTCTCTCACTTTGAGAAAGATAAGTACTACATC
GGGCTTCGACTTGATGCTCGTGTTGACATGTGGAAGAGTAATACAAGCGGTGCAACAAGC
GATGCGAAGCGGTTTGATCTTAACCTCTTACAAGATACAATATTCACCGCGATTTCTGCT
CCAAGATTCCACGAACCGGATTTGGAGCGCTTTCGGGACACAATTGATAACAACCTGAAG
GAGTTACGTTGGCAGCGCCCAGCGCAGTATGCTATTCAACAACTTGAAACCGTTGCATTT
TACAAGGAGCCGCTGGGTAACCCCCGCCACGTTCCAGAGTGGAGTAACGGGCGCATTGCA
AGCAAGGCTTTGCTCGATCAGTATGCCAGATATGTAACTCCAGATCGTGTCGTTGTTGCC
GGTGTCAATATGGAGCACGATGAACTCGTCGCCGAGTATGAAAGCAACCCATATCCGCAT
TCTAGCAACGCCCCGCACCACGCCGCGTTTGCCAAGGGTCTGGGCAAACCTGCTTTTGAC
ATCACGAATGAACACTCCCAGTACAGCGGCGGTGAACTCCACGAACACGAAGACCGGCCA
AAGGAAATGTGTACGAAACCAGATATGGACACCGAGACCGCCATTGCAGTCGGTTATCTT
GCGTTTGGTCGCTCCAAAACGTCCTTGCAGCGCTATGCGGCCACTCTTGTTTACCAACAG
CTGTTCAACATAGTCATCCACGACAGTTTACGCTACGAGCAAGATGTCGTGCTGGACGGT
GTGCGATCGTTTTACCTACCGTATCACTCTGCTGGACTTGTGGGATTCACCGCTATAACC
TCGCCAGAAAATGCTGTGCCGCTGGTTAAAGCGACAATGAAGGGCATTCAGAACGTGAAA
TTTGATAATGCAGCACTATTGGAAGCAGCCAAGCACCGCGCTGCGGTTGAACTAACAAGT
CAGTGCTGGGATTCGTCTCGCGACATATGTGACTATCTGGGCACAAGTTTGTCTCTTGAC
GCCAAGGCTTCCAACTCAACACAATACTTAAATCCCTCAGAGGTGCTAGCGGCTGTTCGC
AACGTCACAGCCAGTGAGTTGAAAGAAGTGAAAGAGTGCATGATGGGCTCAAAACCCTCA
CTGTTTGGGCACGGGGAGATGTTGGCCTTCCCTTCACTCCGTCAGTTGGGAGCGTAA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
Tbg972.11.4600 | 401 S | TAITSPENA | 0.992 | unsp | Tbg972.11.4600 | 445 S | QCWDSSRDI | 0.993 | unsp |


