

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.11.5810 | OTHER | 0.999757 | 0.000166 | 0.000077 |
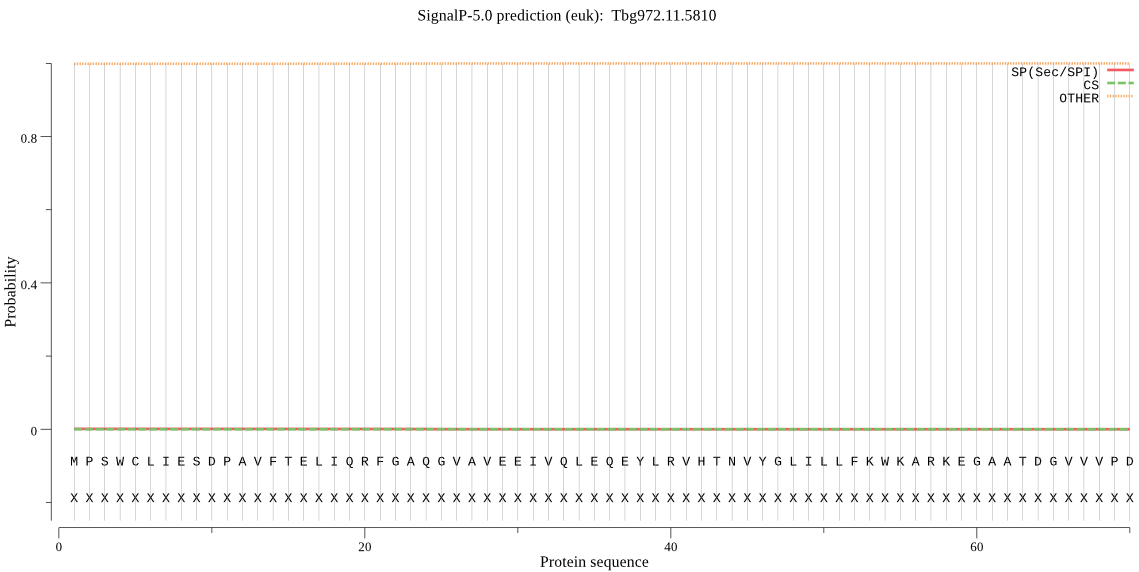
MPSWCLIESDPAVFTELIQRFGAQGVAVEEIVQLEQEYLRVHTNVYGLILLFKWKARKEG AATDGVVVPDAPVFFAQQTVNNACATLSIVNILLNHKESIELGEVLGNFLSFTQDMNPYL RGTQVGECDALREAHNSFAPVELFSLDHSVPDAGDAYHFVSFVYKNSAIWELDGLQEGPI LASDASDENYRDKLMEVVRKRIRDHSAEDTTGAGQGISFSLMAVVDDPLVLLERRIYEAT LQEAPTHYLEEQLNQLTKQRAREKLENQRRRHNYVPMIVELLKALAEKGQLKGILDDALA KKSGQGAKQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.11.5810 | 149 S | SLDHSVPDA | 0.994 | unsp | Tbg972.11.5810 | 206 S | IRDHSAEDT | 0.998 | unsp |