

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
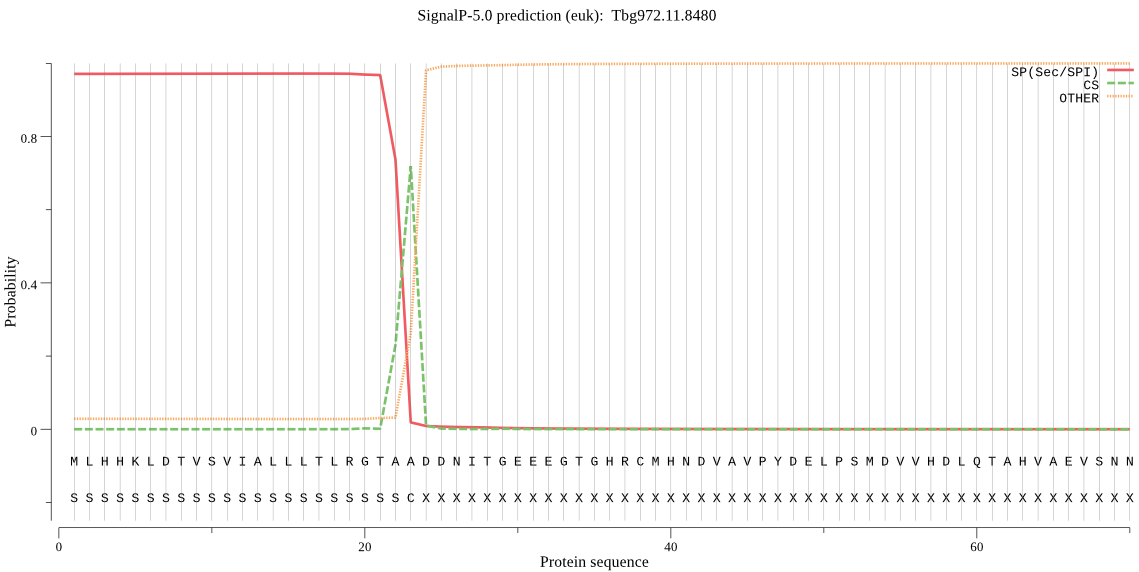
| Tbg972.11.8480 | SP | 0.006957 | 0.992873 | 0.000170 | CS pos: 23-24. TAA-DD. Pr: 0.7995 |
MLHHKLDTVSVIALLLTLRGTAADDNITGEEEGTGHRCMHNDVAVPYDELPSMDVVHDLQ TAHVAEVSNNTEGEGNKSKSVERKNVRFHIKYALGETCKGIGMTVPTYIKGTTKECTEDD VLTKWKLRSVKVMMEAATKFLSSALLVDPLEAVNVPGGKCSGVQVPKMTVPNADYVVFVT INPRPEEETTTVAWAAACRKDTRSGRPVVGHINFIPAAIQRNPSSLAEHVAMHELAHAIG FSDIAETMLRAPNGLGAKGSQRVYRKGLGKAVTLITSPKVLKVAREYYGCPGLDGVEVED AGSEGTRGSHWKKRILFNEALVGSVTSGQLFFSPLTLAYFEDLGFYTANYSTAETGMTWG KGRGCDFLYQKCDSHPREWGEFCFRKEMFVSTCTLDRSSLGACDITTHPEDLPQLYRYFD DPRVGGSSAEMDYCPTVMGFVNAYCTAELGFAFMNVFGNEMGVHSLCYDSDVITSVFPNF PFAARCFPTTCTPSGQLLLRVQGRTVACPRDGKAGLGDTSKLKGVHGKVQCPPSENFCKN SGNGISKLQLASVADEVDGSSNTERIGHSLISPTPHTWNSEDMGSCSSRLACLKDIPPPF PACSLAARKVKECLGNDCPGSAQQWRYANEVGNSCLNPEGMVAMCMDGWRGVNELCGAVD PESKLGRSYRSMLPF
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| Tbg972.11.8480 | 80 S | NKSKSVERK | 0.994 | unsp |