

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
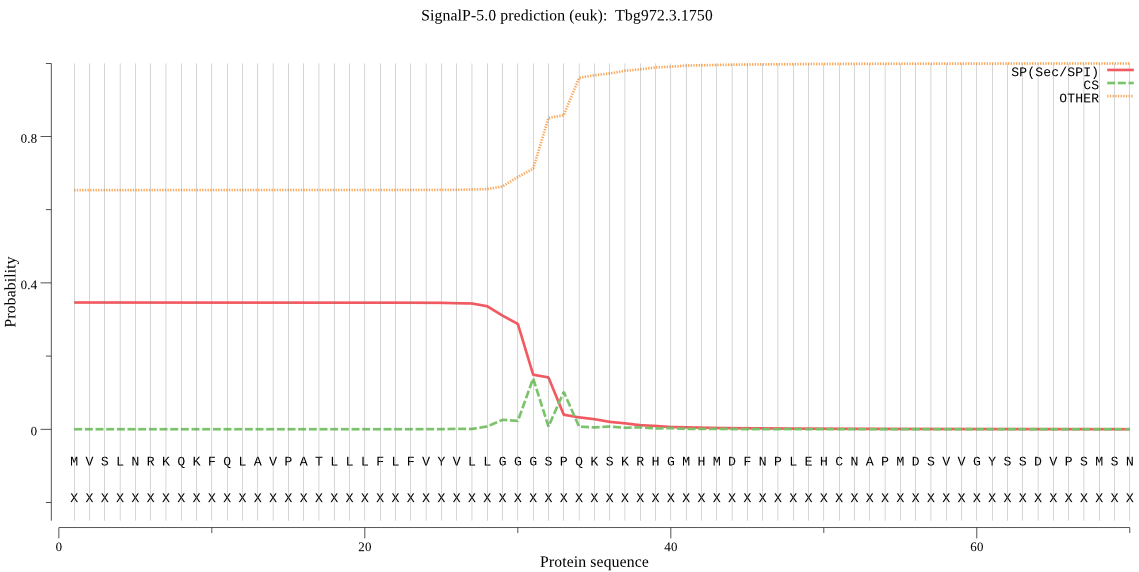
| Tbg972.3.1750 | SP | 0.320622 | 0.678148 | 0.001230 | CS pos: 31-32. GGG-SP. Pr: 0.4902 |
MVSLNRKQKFQLAVPATLLLFLFVYVLLGGGSPQKSKRHGMHMDFNPLEHCNAPMDSVVG YSSDVPSMSNCHRHWLSETYAYTTIGYPHEVMKKMPYGSEWKRAPTGLCWTADEFVARYL LHTRGIFVYFGGSRHDLYWENLKFFGSSGMHRLYERINFENKVEVRTTKRRKRHTPLVGD VVVWNSDYKAYFPRGHVAVVVKVEDDVSAAGGEAALRELKKERRQPSLVYIAEQNFDNKN WEGRNFSRVLKFTWMRGDRASLEDPDGPPLMGHVRVGKLLEDASFFGDL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.3.1750 | 168 T | EVRTTKRRK | 0.99 | unsp | Tbg972.3.1750 | 261 S | GDRASLEDP | 0.998 | unsp |