

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
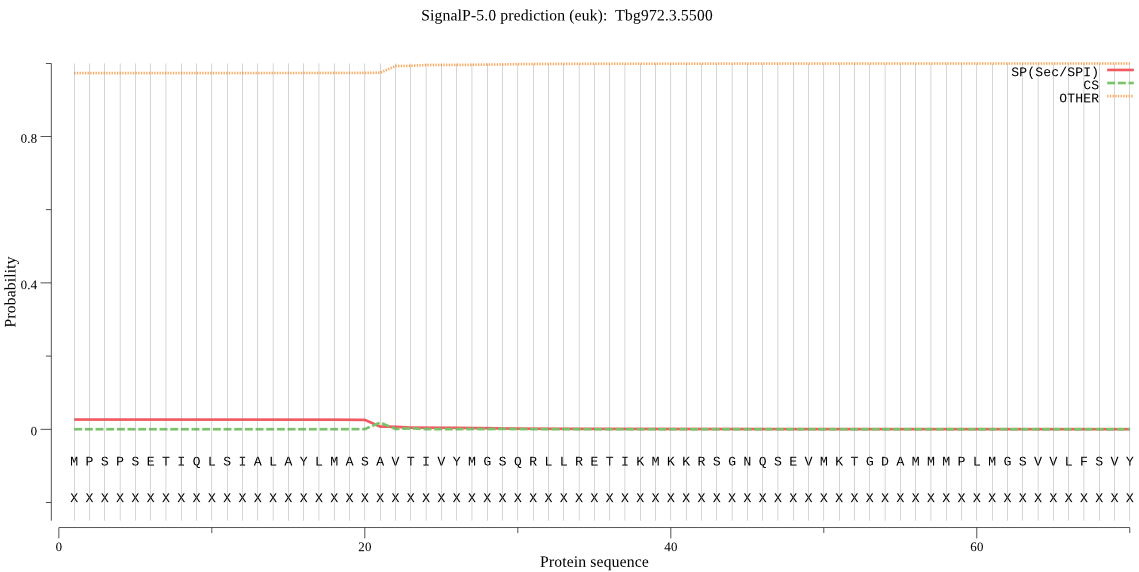
| Tbg972.3.5500 | OTHER | 0.998769 | 0.000909 | 0.000322 |
MPSPSETIQLSIALAYLMASAVTIVYMGSQRLLRETIKMKKRSGNQSEVMKTGDAMMMPL MGSVVLFSVYVVLRFVPREYFNSIVSFYLSIFGVFSLGSFVKTYTRPNVLTGCFCCVAGG LYYITGNWVVNNILATGIAVSAISSIHLGSFKSSFVLLLGLFFYDIFWVFGSDVMLMVAS GVDGPIKMVFPRDIFGGCKSMSLLGLGDLIIPGFFIGQTLVFSSQYVKKGSLYFNVALTA YGLSLVNTMAVMVIFDHGQPALLFIVPWLLVSFSITAVIQGDYKAAWEYTSDAVTEPDNS STDKVKSEEGDQRGTGDEMGLGDFLVKQMKGLFVWDGEEEEEEVCEGTKKND
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.3.5500 | 300 S | EPDNSSTDK | 0.996 | unsp | Tbg972.3.5500 | 307 S | DKVKSEEGD | 0.996 | unsp |