

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
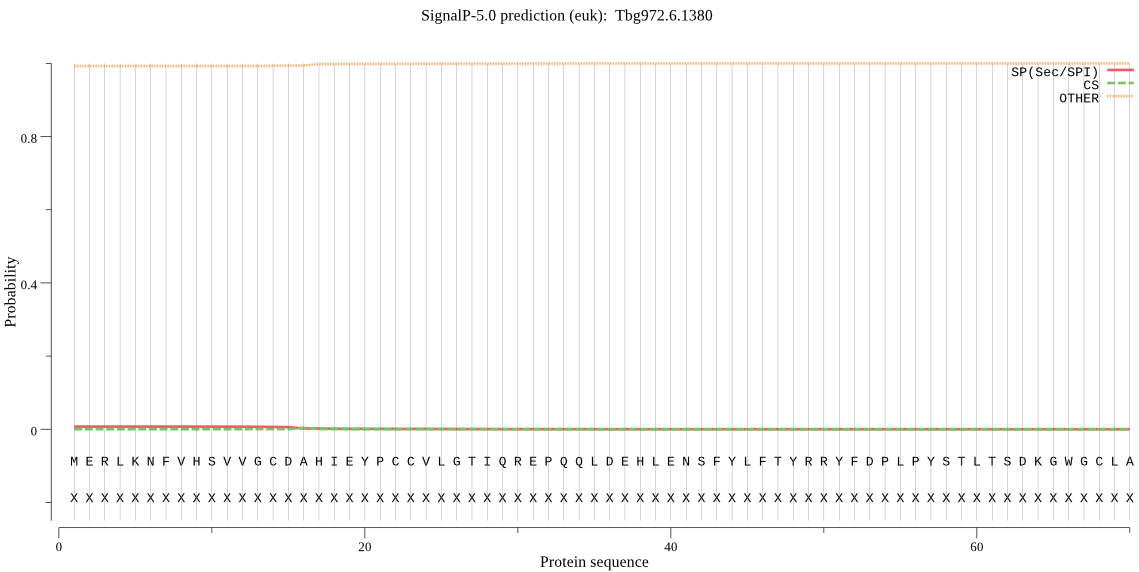
| Tbg972.6.1380 | OTHER | 0.999183 | 0.000778 | 0.000039 |
MERLKNFVHSVVGCDAHIEYPCCVLGTIQREPQQLDEHLENSFYLFTYRRYFDPLPYSTL TSDKGWGCLARATQMLLACSLRRHSAQDCKLQYFADLDDEQVAPFSLHCMVRHILKQGES LRPVYWAPSQGCEAISGCVKRATERGILSSPLSVVITVAGAVPAEEVSCHLKESRNVLIL APLRCGASRCMSQKMFLSLEHLLLAPESVGMVGGVPNRGYYIIGTGAQELLLYLDPHCKT QDALLSGEPGETGVVKPTSSNLRSVPYGQVDTSFFLGFFVDSQSRWESLQKRIEGLSKQK LHPIVSVYRGGPHTPVDLLVMEWPTEP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.6.1380 | 85 S | LRRHSAQDC | 0.998 | unsp | Tbg972.6.1380 | 192 S | SRCMSQKMF | 0.992 | unsp |