

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.6.1510 | OTHER | 0.981680 | 0.003642 | 0.014678 |
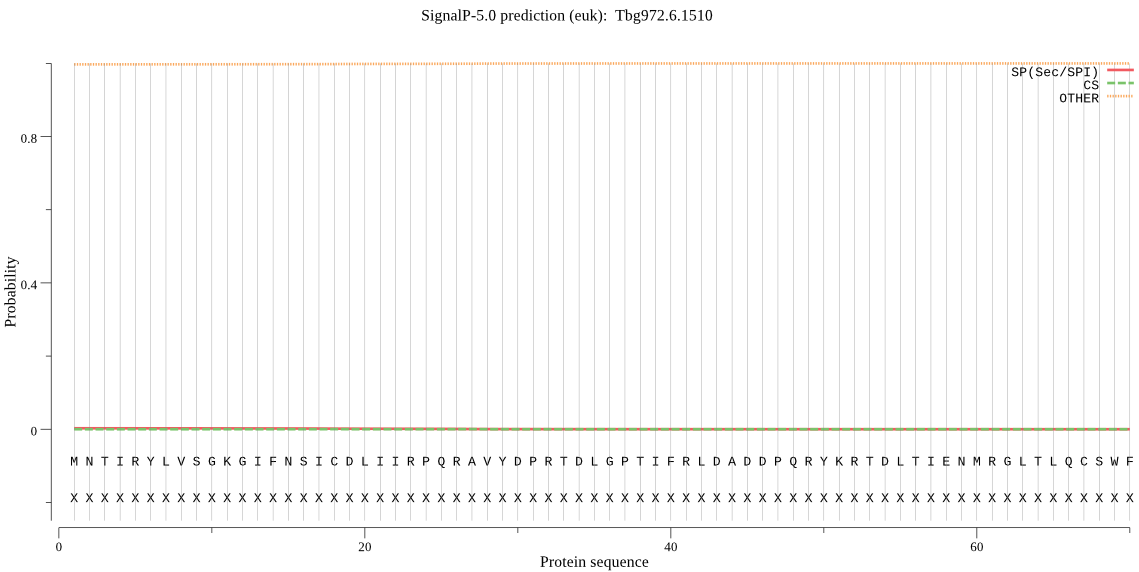
MNTIRYLVSGKGIFNSICDLIIRPQRAVYDPRTDLGPTIFRLDADDPQRYKRTDLTIENM RGLTLQCSWFRTLSNEKQPCIVYIHGNCGSRYDALEALFLLKEGYSLFCFDAAGSGLSDG EYISLGFYERQDLAAVVDYLEDQEEVDGIGLWGRSMGAVTSIMYASKDNSIKCIVCDSPF STLRSLVNDLVKQHGSKRFPSSLINKIVNRMRKRIAARAAFNIDDLDTLKYASECTVPAF IFHGREDDFVFPRNSIDVSNYFMGPCLHHLVDGGHNDERGEDVRNTIKGFFSLYLVLKRE NGRPVAHLMQDATNRVPREYIPRVDSGDVSSSPERDGDDNDNNSGVGGSGDVSVDDGGNS SNSDEVHGLEGSGDEEEPIDEAPSRPLSASLAPVRVTKPWFYDPNDLLLERKD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.6.1510 | 353 S | SGDVSVDDG | 0.996 | unsp | Tbg972.6.1510 | 353 S | SGDVSVDDG | 0.996 | unsp | Tbg972.6.1510 | 353 S | SGDVSVDDG | 0.996 | unsp | Tbg972.6.1510 | 170 S | SKDNSIKCI | 0.992 | unsp | Tbg972.6.1510 | 332 S | DVSSSPERD | 0.997 | unsp |