

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.6.220 | SP | 0.009178 | 0.990399 | 0.000423 | CS pos: 22-23. VNA-AL. Pr: 0.6997 |
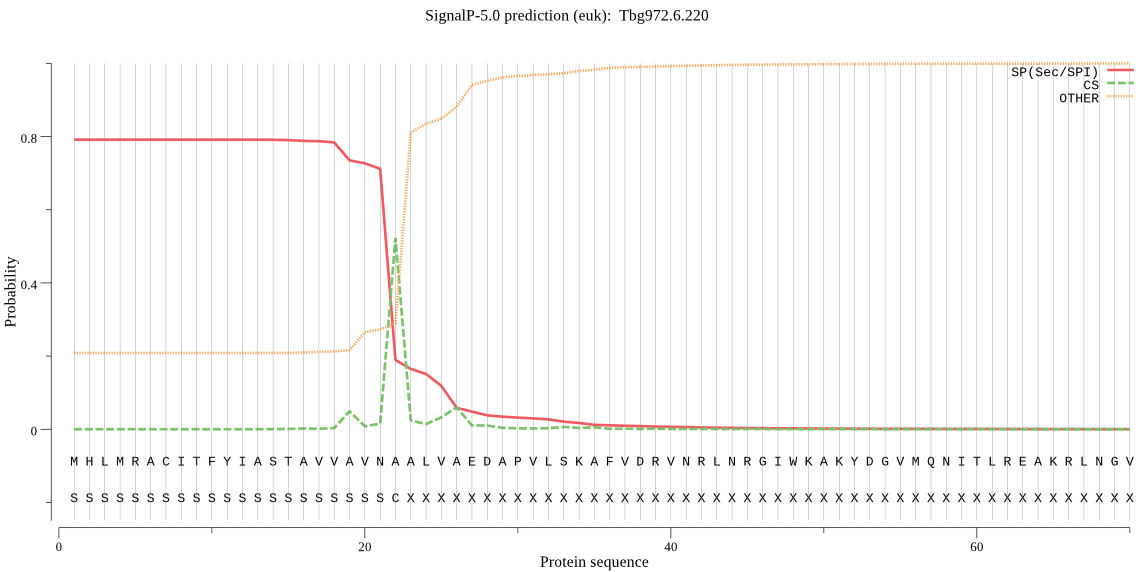
MHLMRACITFYIASTAVVAVNAALVAEDAPVLSKAFVDRVNRLNRGIWKAKYDGVMQNIT LREAKRLNGVIKKNNNASILPKRRFTEEEARAPLPSSFDSAEAWPNCPTIPQIADQSACG SCWAVAAASAMSDRFCTMGGVQDVHISAGDLLACCSDCGDGCNGGDPDRAWAYFSSTGLV SDYCQPYPFPHCSHHSKSKNGYPPCSQFNFDTPKCNYTCDDPTIPVVNYRSWTSYALQGE DDYMRELFFRGPFEVAFDVYEDFIAYNSGVYHHVSGQYLGGHAVRLVGWGTSNGVPYWKI ANSWNTEWGMDGYFLIRRGSSECGIEDGGSAGIPLAPNTA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.6.220 | 321 S | RRGSSECGI | 0.994 | unsp | Tbg972.6.220 | 321 S | RRGSSECGI | 0.994 | unsp | Tbg972.6.220 | 321 S | RRGSSECGI | 0.994 | unsp | Tbg972.6.220 | 86 T | KRRFTEEEA | 0.99 | unsp | Tbg972.6.220 | 320 S | IRRGSSECG | 0.996 | unsp |