

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.6.735 | SP | 0.026565 | 0.972351 | 0.001084 | CS pos: 29-30. ALG-SL. Pr: 0.7726 |
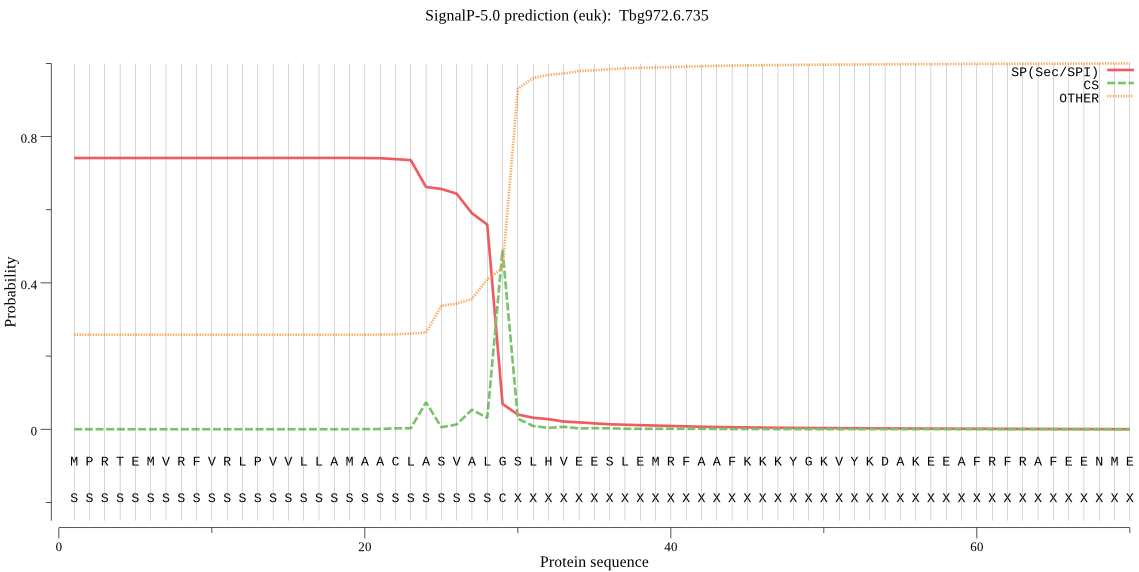
MPRTEMVRFVRLPVVLLAMAACLASVALGSLHVEESLEMRFAAFKKKYGKVYKDAKEEAF RFRAFEENMEQAKIQAAANPYATFGVTPFSDMTREEFRARYRNGASYFAAAQKRLRKTVN VTTGRAPAAVDWREKGAVTPVKDQGQCGSCWAFSTIGNIEGQWQVAGNPLVSLSEQMLVS CDTIDFGCGGGLMDNAFNWIVNSNGGNVFTEASYPYVSGNGEQPQCQMNGHEIGAAITDH VDLPQDEDAIAAYLAENGPLAIAVDATSFMDYNGGILTSCTSEQLDHGVLLVGYNDNSNP PYWIIKNSWSNMWGEDGYIRIEKGTNQCLMNQAVSSAVVGGPTPPPPPPPPPSATFTQDF CEGKGCTKGCSHATFPTGECVQTTGVGSVIATCGASNLTQIIYPLSRSCSGLSVPITVPL DKCIPILIGSVEYHCSTNPPTKAARLVPHQ