

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.7.4990 | OTHER | 0.999839 | 0.000104 | 0.000057 |
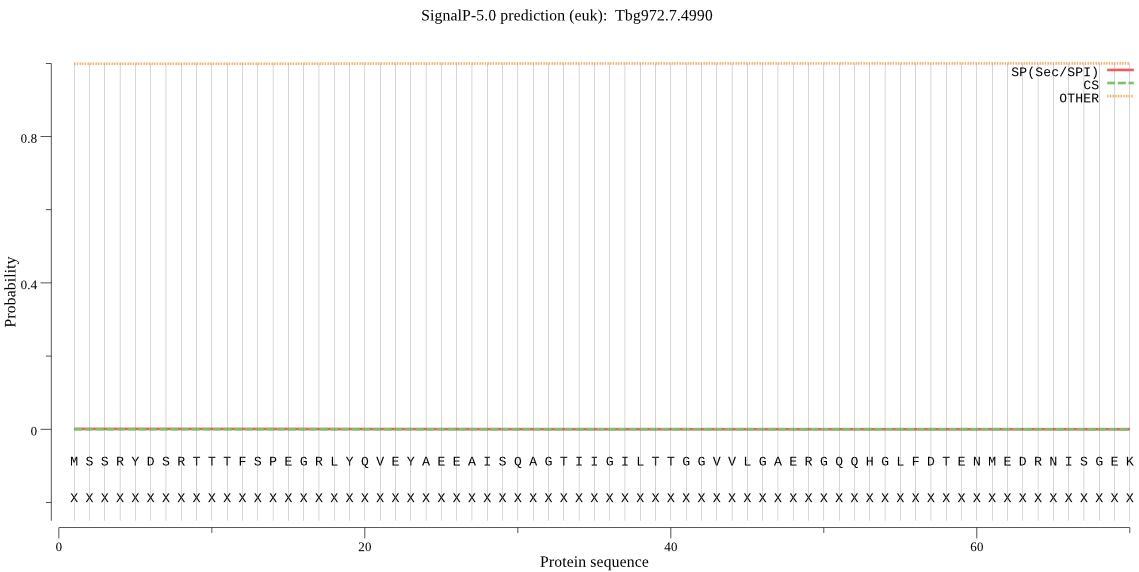
MSSRYDSRTTTFSPEGRLYQVEYAEEAISQAGTIIGILTTGGVVLGAERGQQHGLFDTEN MEDRNISGEKVYKISNHLGCSVAGVTSDAYALINYARLSAARHFYSFQEPIAAEDLCRMV CDEKQLYTQYGGVRPFGVSFLFAGWDRHYGYQLYHTDASGNYSAWRAYAVGQNDQVAQSL LKRDWKPELTLEEGIVLCLRVLGKTMDAVKLTPERLEVAVLQRVQAPLTQKLLDPYGVNP KMVPSFKILTDEELKPHVDEANRQREAEEAEEAEKEKKNEKRLASP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.7.4990 | 7 S | SRYDSRTTT | 0.995 | unsp | Tbg972.7.4990 | 13 S | TTTFSPEGR | 0.99 | unsp |