

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.7.7240 | OTHER | 0.957833 | 0.000775 | 0.041392 |
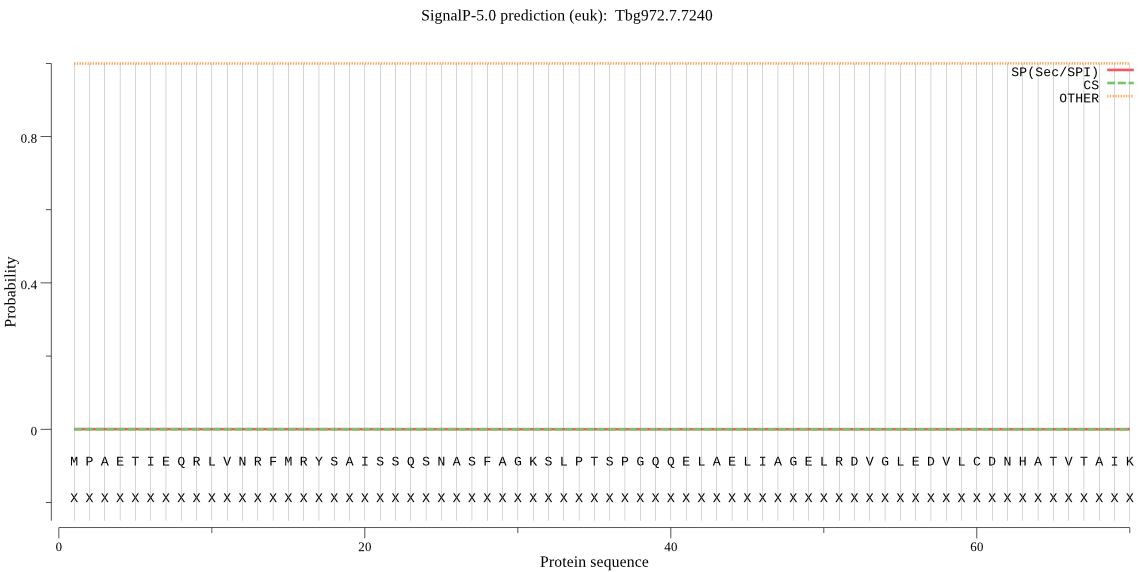
MPAETIEQRLVNRFMRYSAISSQSNASFAGKSLPTSPGQQELAELIAGELRDVGLEDVLC DNHATVTAIKRGTVKDCPTIGFIVHLDTVDVGLSPVVRAQKLHYDGGDVCLNKEKDIWLR VTEHPEIEAYVGQDILFGDGTSVLGADDKAAITVVMEMITGLQPNDEHGDIVVCCVPDEE IGLLGPKNLDLKARFDVDFAYTIDCCELGEVVYENFNAAAARITFTGVTAHPMSAKGVLV NPLLMAMDYIAQFDRNETPECTAGRDGYWWFSDIYANQNEAVLDVKIREFDAVRFEARKQ QLLSVAESIRLCYPTGGVELSISEQYRNIANFLGDDRSAVDLLFDALRSVGVQPKVTPMR GGTDGAVLSSRGVVTPNYFTGAHNFHSKFEFLPIPSFVKSYEVTRQLALLATKLPKLRKR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.7.7240 | 36 S | SLPTSPGQQ | 0.996 | unsp | Tbg972.7.7240 | 400 S | SFVKSYEVT | 0.993 | unsp |