

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.8.150 | OTHER | 0.999938 | 0.000052 | 0.000010 |
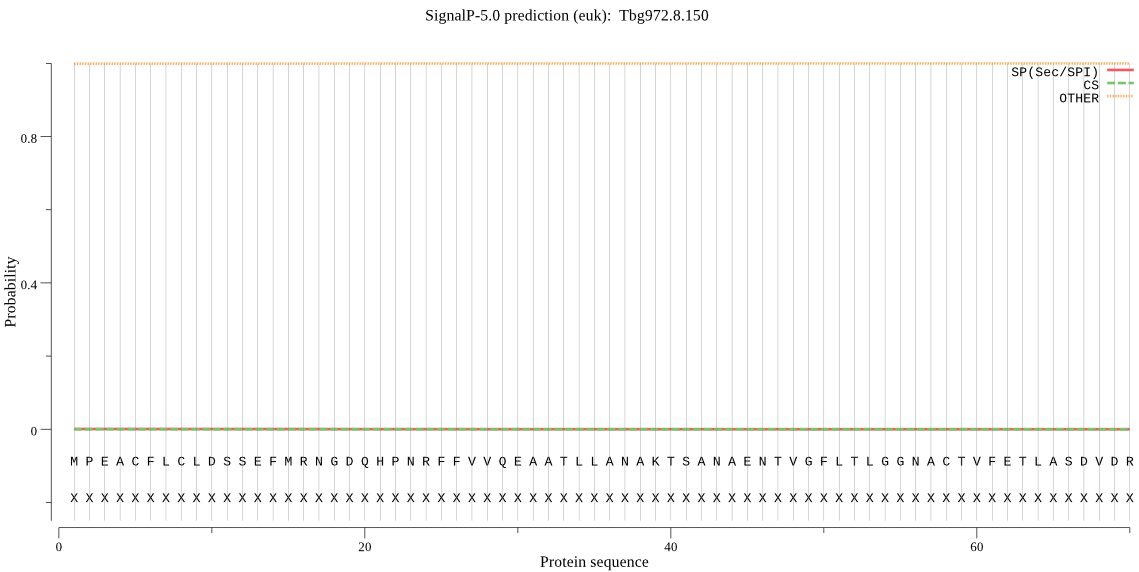
MPEACFLCLDSSEFMRNGDQHPNRFFVVQEAATLLANAKTSANAENTVGFLTLGGNACTV FETLASDVDRVMSTMSKISISGKQCHFSKGLQIACLALSHRTNPRAEKRIVAFIGTPLGE TDGELEKLAKKLRKESVAVDVVSVGVPSNVPRLTAFVERLSNNGNSRFLSVPAKVPLIDS LMSSAIFMGEDSSVSGGGFNGSSAGFSVDPTMDPEMVLAIQMSLEEEERRQAAAAALNSN AAESETVERTDINMDEEALSLENMTEEEMLRRALLLSLQEANKGQAAAGSNATNTADDEA FANELEKELERQSKEETLQGKHDDQNEEKED
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| Tbg972.8.150 | 313 S | LERQSKEET | 0.998 | unsp |