

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
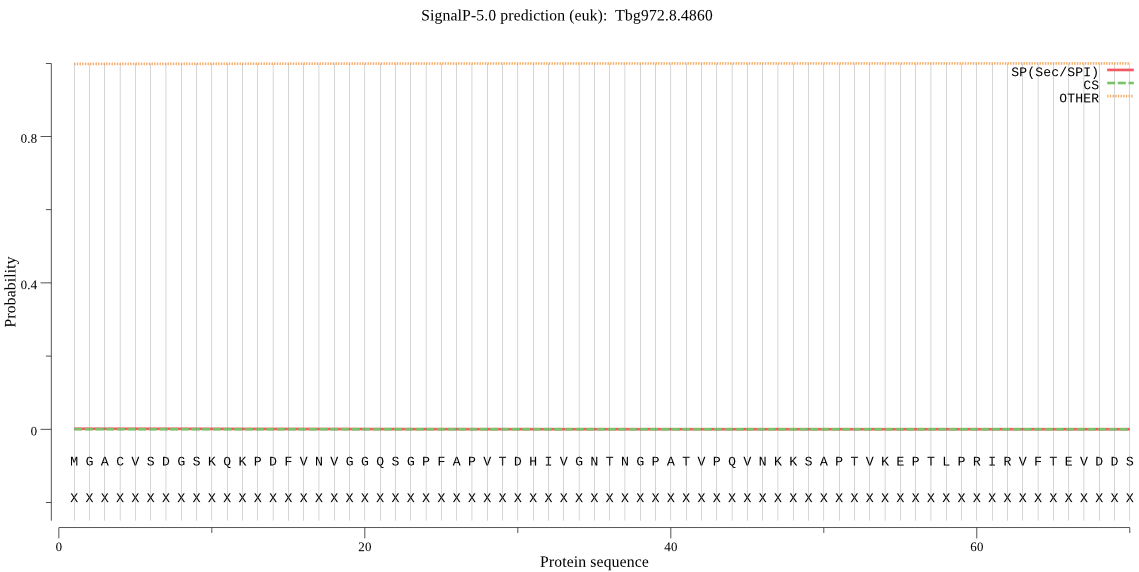
| Tbg972.8.4860 | OTHER | 0.999844 | 0.000147 | 0.000010 |
MGACVSDGSKQKPDFVNVGGQSGPFAPVTDHIVGNTNGPATVPQVNKKSAPTVKEPTLPR IRVFTEVDDSEANNIDLVPGSGCSVVKGKYRMTPIVAFKPFAARRLRERAVKVIRGSYYS EEAHEEQPQACAVTSCPFGDEEKEGASGTKKCNQPEFSFALDTSTDTEEGLSSGCDTSAS PRDRSLAMCGGEKGCRNEPADECTPNRRGSLPSGVGGDGPQSHKKGIDVVDKLNTPGTAR SVNGITTEPKSTPRLSREDLILLGSKRLKKRVEELRLIEHQMKDDGNCQFRAISHQIFGS QEYHELVRVHVVTYMKSVRDSFDCFLGTTEDADHYYADMLKNGTWGDELTLRAASDSLFI NIHILSSEEQNYYITYNPSPDAPTPPAFLVDVATMRQDRQEAFSTSVASVCASPSLSARG SWNGGGGKKSLATPRGPVDQLGLAGINRNFRPIHEEVNVVELQREVQRKLAASTIRNTTL PVPEPLLPFKAVIEQNVVVPPTAAPQPKPTFVDAPKSPVKTVRIELSPREVSFNAKNTSV IFVCARKKPRRKRTSRGRSKTRGGRIVGLPKRHPIMRPISFSSSPITARRRPRCRSALVR NLSPRRSKKLRSKSSAMPASLQLPPFASPSESPVNPRLSSPEPAPVIELPGTASFILVSP RSSDSDMVKRLSLGSASVPHSSPPPRSAVCEGSASPKVSVSPSEKEELESCFLSEAPNAP IDIFLSYLSPVHYNALSVDTISRPVE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.8.4860 | 173 S | EGLSSGCDT | 0.994 | unsp | Tbg972.8.4860 | 173 S | EGLSSGCDT | 0.994 | unsp | Tbg972.8.4860 | 173 S | EGLSSGCDT | 0.994 | unsp | Tbg972.8.4860 | 180 S | DTSASPRDR | 0.995 | unsp | Tbg972.8.4860 | 210 S | NRRGSLPSG | 0.995 | unsp | Tbg972.8.4860 | 222 S | DGPQSHKKG | 0.99 | unsp | Tbg972.8.4860 | 251 S | TEPKSTPRL | 0.991 | unsp | Tbg972.8.4860 | 256 S | TPRLSREDL | 0.998 | unsp | Tbg972.8.4860 | 555 S | RKRTSRGRS | 0.998 | unsp | Tbg972.8.4860 | 559 S | SRGRSKTRG | 0.997 | unsp | Tbg972.8.4860 | 580 S | MRPISFSSS | 0.991 | unsp | Tbg972.8.4860 | 584 S | SFSSSPITA | 0.99 | unsp | Tbg972.8.4860 | 603 S | VRNLSPRRS | 0.997 | unsp | Tbg972.8.4860 | 632 S | SPSESPVNP | 0.993 | unsp | Tbg972.8.4860 | 639 S | NPRLSSPEP | 0.993 | unsp | Tbg972.8.4860 | 640 S | PRLSSPEPA | 0.995 | unsp | Tbg972.8.4860 | 663 S | SPRSSDSDM | 0.997 | unsp | Tbg972.8.4860 | 672 S | VKRLSLGSA | 0.993 | unsp | Tbg972.8.4860 | 701 S | KVSVSPSEK | 0.997 | unsp | Tbg972.8.4860 | 703 S | SVSPSEKEE | 0.996 | unsp | Tbg972.8.4860 | 117 S | VIRGSYYSE | 0.994 | unsp | Tbg972.8.4860 | 120 S | GSYYSEEAH | 0.993 | unsp |