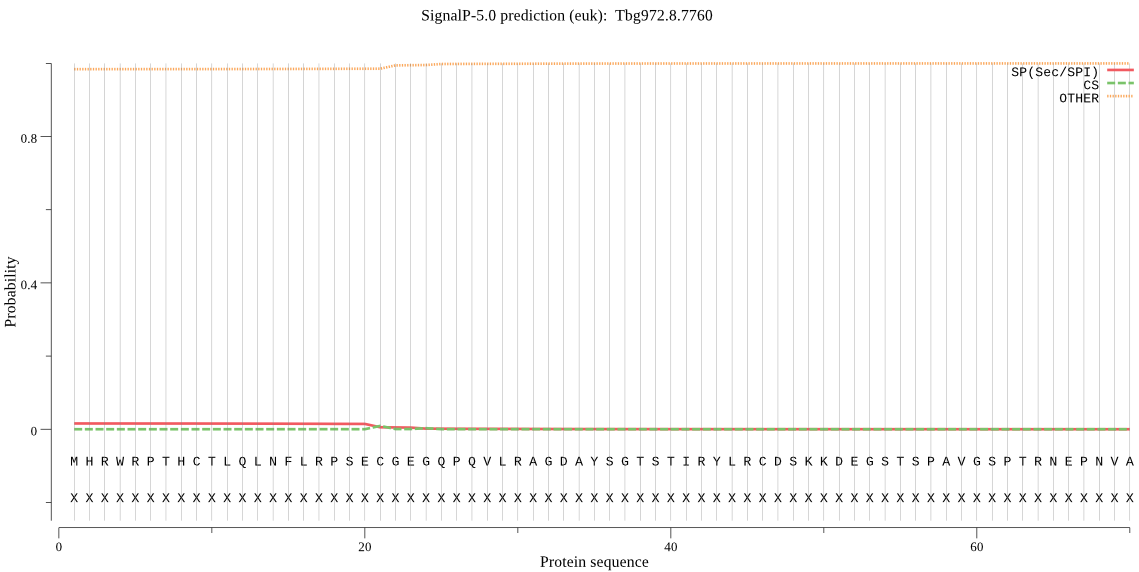
Fasta :-
MHRWRPTHCTLQLNFLRPSECGEGQPQVLRAGDAYSGTSTIRYLRCDSKKDEGSTSPAVG
SPTRNEPNVAIPRKEDGLHFHAITNIPHSLPKALPLTLQSMKVQMASQRSGAVSLKVLKV
VDESNSISDGNKNNSGTGPTGASRSDKHVKNCEARKREKQSLKKKQRCKNSDEHNSMGGD
GSCEQQADDPGSKYVYFEGGADMPAGSTVLLSIAFTGRVQAWDQGGIYAGEQKEGGAGSR
VLLTHFEVALARLAFPCPDDPQEYRIVWQLQLLQLPAEYNMVVSNTTEITKKAVGSRGMQ
YSFAPIGPLPAYVIAFAAFAGTVEVLEDLQLRGLPAVEYIEQTTSGKSLSAHDGRGNHAM
RTVTMRVVTHATSGVTPPVLAQVAHTARDAIRLLEDFFDSPIPLQQLPFCSSHSSNDHWG
SDYYCCEWQQYGGGETLTIVVAPTMPYISGMEHHGCIFLNESIYSLTQSVGGTGARGASR
SNAPGSETVVEVERVELIVHELVHHWVGNALGMPFVLKEGVCQLMEQCLGDVIMGRPMRK
VRPAATGANTTASNINTDSTKTRADDANLVIVDAEKGKEFTFHSYQKALNTLRNVVSVMG
FNVFKERMQRMYASEVLEDARSKTSSLPPYVSATKFMAYMNPQDAALYCT