

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.8.8340 | OTHER | 0.968814 | 0.031100 | 0.000085 |
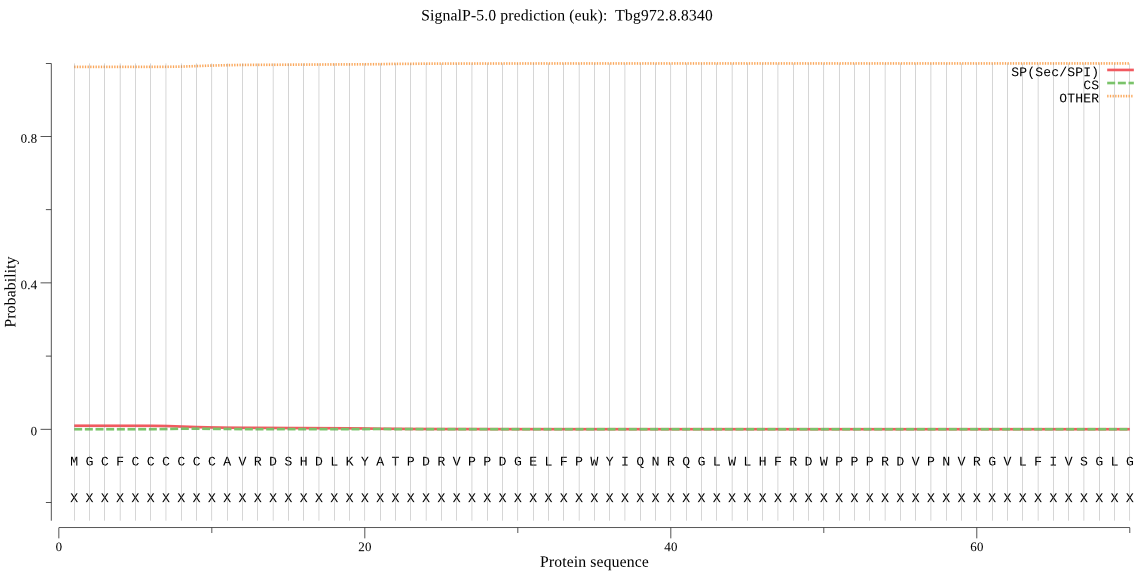
MGCFCCCCCCAVRDSHDLKYATPDRVPPDGELFPWYIQNRQGLWLHFRDWPPPRDVPNVR GVLFIVSGLGEHTARYGGVGRYFSREGFHVFCMDNQGAGASEGARLYVSDFDDFIVDFFL FKRHVFSLYPEYEALPRFLLGHSMGGLIATHVSLRDPTGFTGFIFSGPALKPHPKLASCF KQCCVGLMSSCVPKFGVGSIDPKSVSTNRQVVELLEQDPLNFDAKLTARWGKTMLDAMES VWTQVERATYPVLILHGAKDALCPISGSRKFLESVPTTDKQLIEYPGLGHEVLTEVRWRE VLGDILKFINAHCK