

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
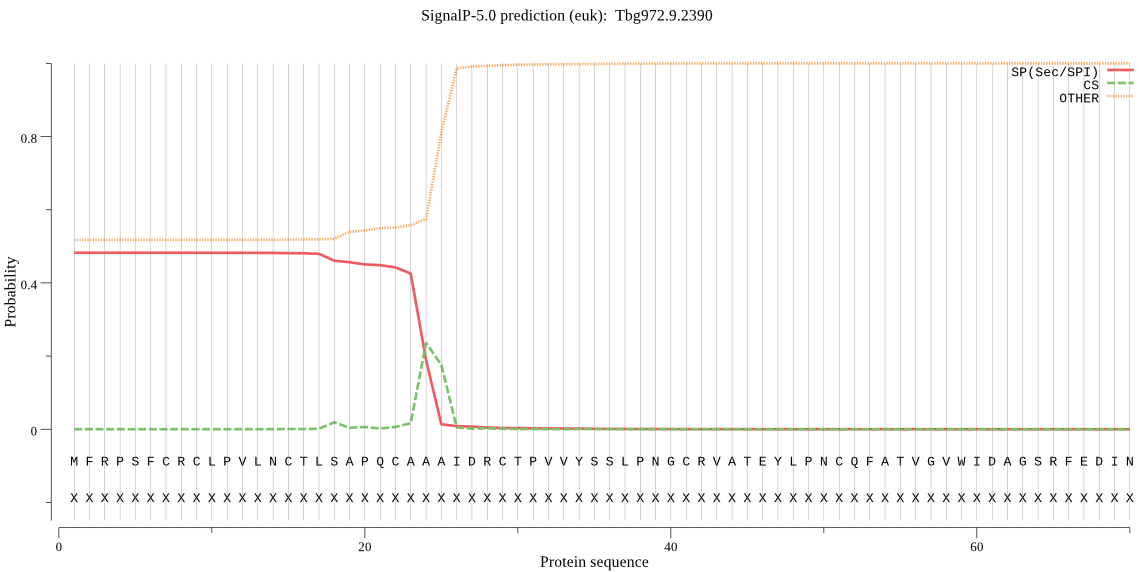
| Tbg972.9.2390 | SP | 0.259847 | 0.463046 | 0.277107 | CS pos: 24-25. CAA-AI. Pr: 0.5199 |
MFRPSFCRCLPVLNCTLSAPQCAAAIDRCTPVVYSSLPNGCRVATEYLPNCQFATVGVWI DAGSRFEDINNNGVAHFLEHMNFKGTAKYSKRAVEDLFEHRGAHFNAYTSRDRTAYYVKA FKYDVEKMIDVVSDLLQNGRYDPSDVELERPTILAEMREVEELVDEVLMDNLHQAAYDPA HCGLPLTILGPVENISSRINRDMIQEFVRVHYTGPRMSFISSGGIHPDEAHRLAEKFFGN LPAANNSPLLQSQYRGGYTVMWNEQMATANTAFAYPICGAIHDDSYALQLVHNVIGQVRE GQHDQFAHQRLNPRLPWEKLSNLVQLRPFYTPYKETSLLGYQLVTMRTAVADANGGVQRD ESQTVLLDHMLKLFNELSTKAVDAALLEEAKSEYKSSVMMMRDSTTNSAEDLGRQMIHLG RRVPLREVFERVDAVTPAVFRDTLAKYVQAVQPTVSYIGSASAVPRFDALTQVKHIL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.9.2390 | 404 S | MMRDSTTNS | 0.991 | unsp | Tbg972.9.2390 | 408 S | STTNSAEDL | 0.994 | unsp |