

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.9.7990 | OTHER | 0.999929 | 0.000024 | 0.000047 |
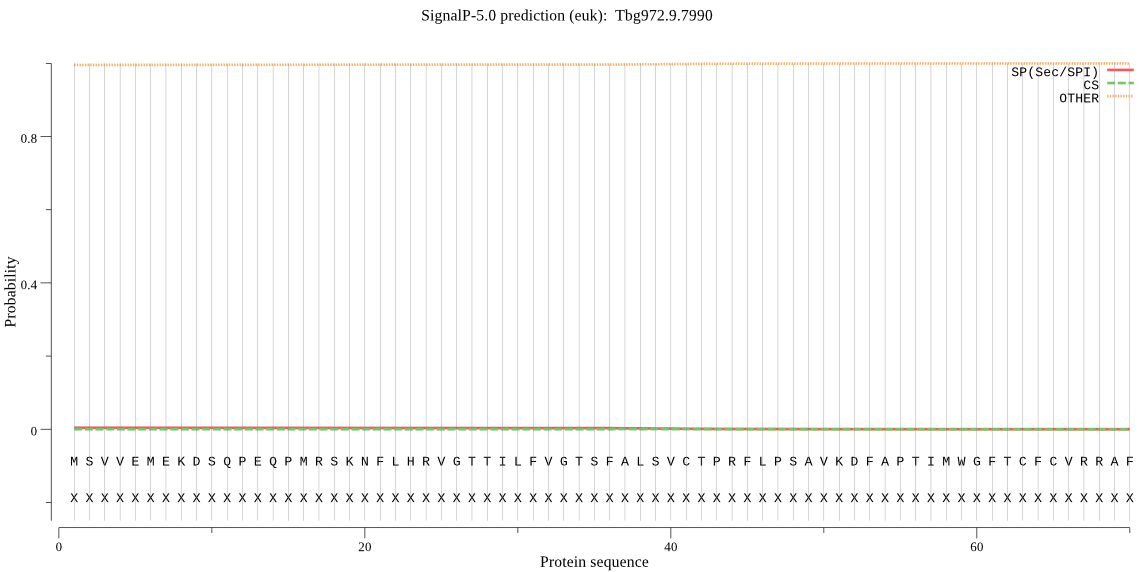
MSVVEMEKDSQPEQPMRSKNFLHRVGTTILFVGTSFALSVCTPRFLPSAVKDFAPTIMWG FTCFCVRRAFKMMMTLVLITHLLASGASLWSCGFSNRVANRALIFPSFASLAALLATWRL PTMPLERVIGVSLVQTWSISCGLILFAVFPATLISGSNWDQAKRDARFDQVIERATDQRV ERFTVQSTGGVQLDAVAVMPEDPTEHWVAYFGGNAEIMETSTEDMAFSNHLVHANWLFHN PRGVGRSSGYVCWANDLVDDAEAVVWEAVRRYNINPKNLILWGHSIGGGVAAALAGRYSD FPFPVLLDRTFSRLSDAAVSFSPFPATVTRGAIKVTTGDLNVVKSCRSLKGRNVLVIFHR SDEIIRFNISSIARPTAVKKSGLNKSSFLELQSTFVPSPHNTPLDYFPEVTQIQRRIIQL YE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.9.7990 | 398 S | TFVPSPHNT | 0.992 | unsp | Tbg972.9.7990 | 398 S | TFVPSPHNT | 0.992 | unsp | Tbg972.9.7990 | 398 S | TFVPSPHNT | 0.992 | unsp | Tbg972.9.7990 | 10 S | MEKDSQPEQ | 0.992 | unsp | Tbg972.9.7990 | 221 S | IMETSTEDM | 0.993 | unsp |