

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
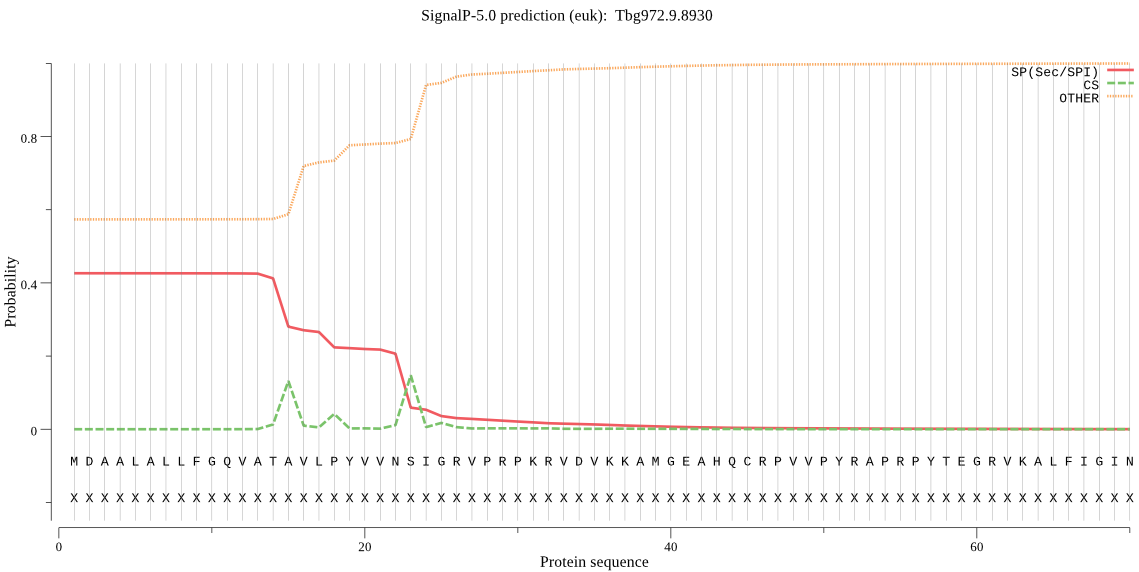
| Tbg972.9.8930 | SP | 0.136439 | 0.855081 | 0.008480 | CS pos: 23-24. VNS-IG. Pr: 0.3937 |
MDAALALLFGQVATAVLPYVVNSIGRVPRPKRVDVKKAMGEAHQCRPVVPYRAPRPYTEG RVKALFIGINYTGSSAQLGGCVNDVMHMLQTLQRIEFPISECCILVDDRRFPNFTAMPTR ENIIKYMAWLVYDVRPGDVLFFHFSGHGAETKGGRDSNEKMDQCLVPLDYDKAGAILDDD LFELMIKGLPAGVRMTAVFDCCHSASLLDLPFAFVAGRNVSSNQRHEMRMVRKDNYSRGD VVMFSGCEDSGTSADVTNTSSFGNGTVAAGGAATQAFTWALLNTTGYSYIDIFMKTREVL RQKGYKQVPQLSSSKPVDLYKQFSLFGPLTMNASLVQHLPQEYVQPWAPHPAYQQPHEAT LPASVSQPHSQPVMGIPVASTSNGKSNPGVSDGGRASGEVYPPTQYPSSHPAPQQQAYYQ PPQQAYYQPPQQAYYQPPQQAYYQPPQQAYYQPEPHHQPAPPPPPKKENKPARPGYPMSY CMKFSQGKPGRK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.9.8930 | 397 S | GGRASGEVY | 0.991 | unsp | Tbg972.9.8930 | 397 S | GGRASGEVY | 0.991 | unsp | Tbg972.9.8930 | 397 S | GGRASGEVY | 0.991 | unsp | Tbg972.9.8930 | 157 S | GGRDSNEKM | 0.995 | unsp | Tbg972.9.8930 | 237 S | KDNYSRGDV | 0.993 | unsp |