

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
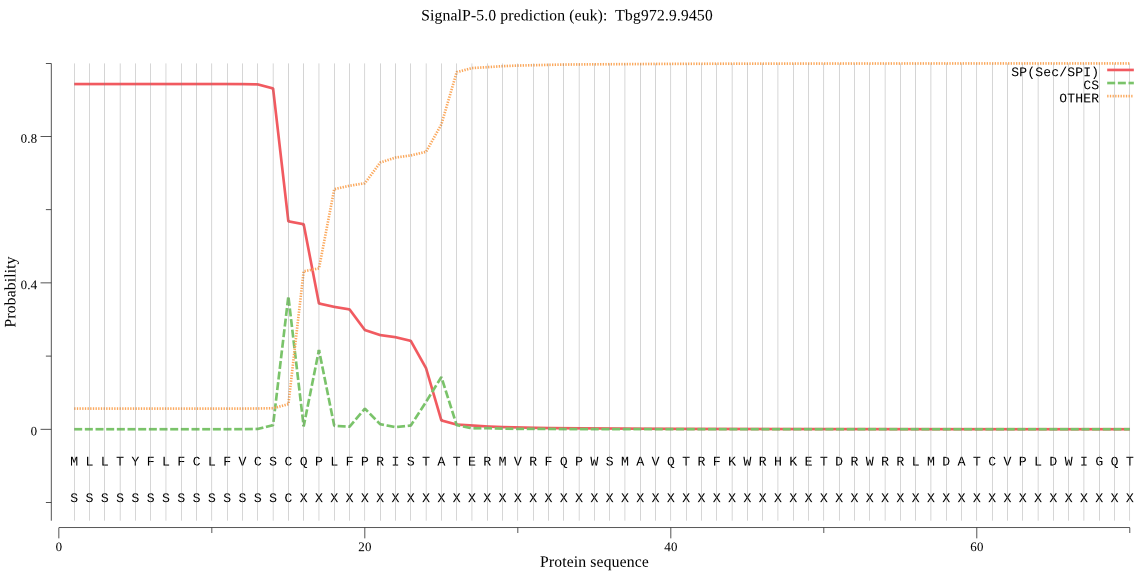
| Tbg972.9.9450 | SP | 0.017565 | 0.972503 | 0.009932 | CS pos: 15-16. CSC-QP. Pr: 0.2992 |
MLLTYFLFCLFVCSCQPLFPRISTATERMVRFQPWSMAVQTRFKWRHKETDRWRRLMDAT CVPLDWIGQTRGPSFTQYSGHWTHLLTCAHVITPWDYPNFYPPKGPTRFVSRITLADTMT QVRLVSLQGNAVYKHFTSNQHVFVHSNPRLDLCVLHPEQNLRRSGEMKMLWMQNEGYVTR PRLELVKPLEVGDHVWVYGMTAHESLFDEEGGPEPLMIPTGIRARVHALTREHFFIDTKS IEGAERGCVGMGMCGSAVMRNGRCVGMLTATVHEESDCKELAGTAMCTYASDIFEFLLEV ERQMKALPPSLNRQETMFQQRRRGEGSEEYEKQRETEGKEWDLDHTRLARHIPVPVSLWH MEEKWITEEDYMTNKVFGRGGPFNQETQENILGYDMNSSKSYGERPGDIASVSTATQDGK PTMQGERKDYSPTGIYADHEDFKTSDVWDYNVSSELRSLFDKGVDAKDAHSLNMMRKSLE NIRAQRAMEKMKETVMKAPGHSEDPLNVGSMGRYDVNMEESSAQAFTPDTDDEVRAAARK EEKYHEELRRRHGNKARPFGDEDLRGLWEPH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.9.9450 | 478 S | MMRKSLENI | 0.991 | unsp | Tbg972.9.9450 | 478 S | MMRKSLENI | 0.991 | unsp | Tbg972.9.9450 | 478 S | MMRKSLENI | 0.991 | unsp | Tbg972.9.9450 | 164 S | NLRRSGEMK | 0.992 | unsp | Tbg972.9.9450 | 401 S | NSSKSYGER | 0.996 | unsp |