

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
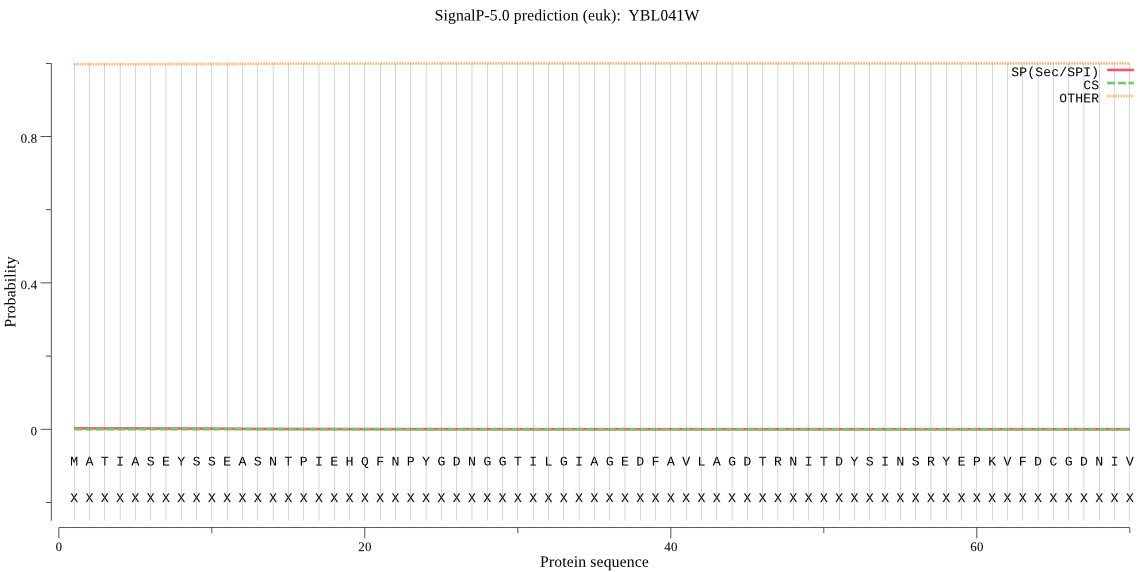
| YBL041W | OTHER | 0.993727 | 0.006258 | 0.000015 |
MATIASEYSSEASNTPIEHQFNPYGDNGGTILGIAGEDFAVLAGDTRNITDYSINSRYEP KVFDCGDNIVMSANGFAADGDALVKRFKNSVKWYHFDHNDKKLSINSAARNIQHLLYGKR FFPYYVHTIIAGLDEDGKGAVYSFDPVGSYEREQCRAGGAAASLIMPFLDNQVNFKNQYE PGTNGKVKKPLKYLSVEEVIKLVRDSFTSATERHIQVGDGLEILIVTKDGVRKEFYELKR D
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| YBL041W | T | 3 | 0.650 | 0.147 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| YBL041W | T | 3 | 0.650 | 0.147 |