

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
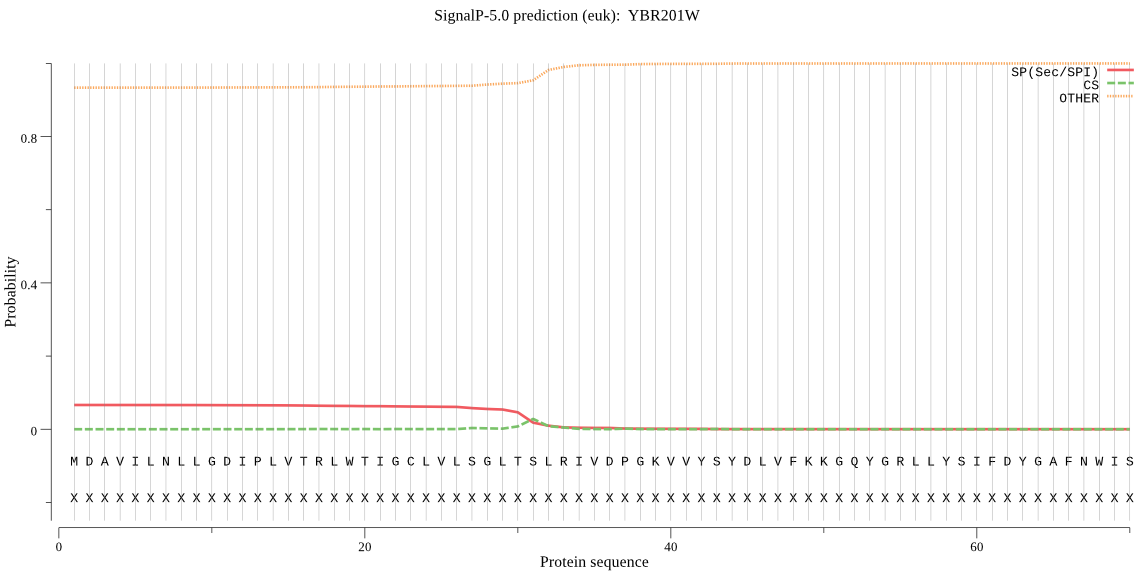
| YBR201W | OTHER | 0.969673 | 0.028218 | 0.002109 |
MDAVILNLLGDIPLVTRLWTIGCLVLSGLTSLRIVDPGKVVYSYDLVFKKGQYGRLLYSI FDYGAFNWISMINIFVSANHLSTLENSFNLRRKFCWIIFLLLVILVKMTSIEQPAASLGV LLHENLVYYELKKNGNQMNVRFFGAIDVSPSIFPIYMNAVMYFVYKRSWLEIAMNFMPGH VIYYMDDIIGKIYGIDLCKSPYDWFRNTETP
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| YBR201W | T | 210 | 0.523 | 0.029 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| YBR201W | T | 210 | 0.523 | 0.029 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| YBR201W | 200 S | DLCKSPYDW | 0.997 | unsp |