

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
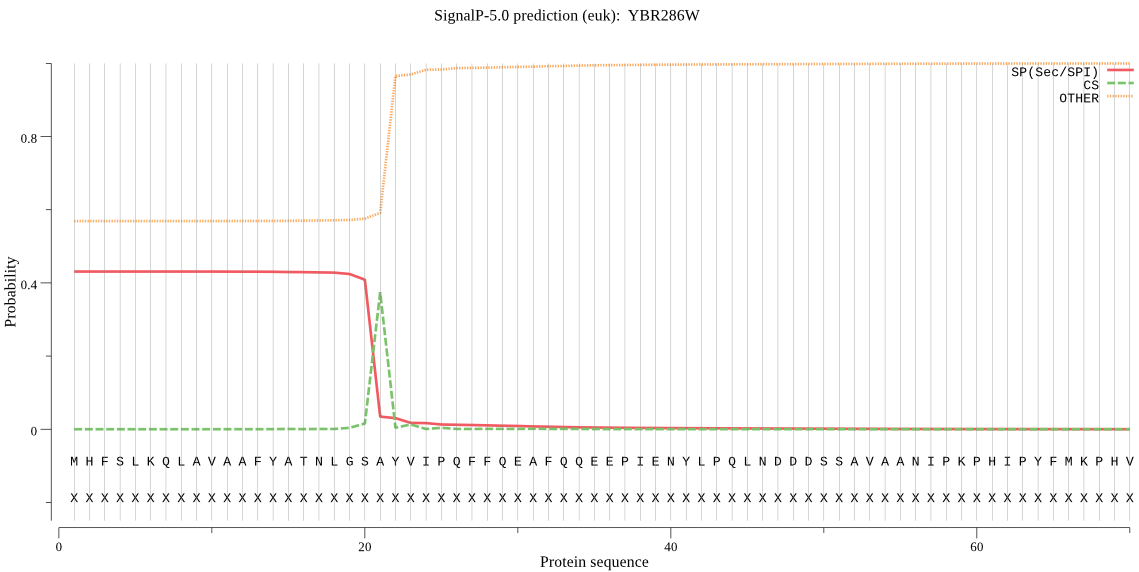
| YBR286W | SP | 0.116770 | 0.869252 | 0.013977 | CS pos: 21-22. GSA-YV. Pr: 0.8799 |
MHFSLKQLAVAAFYATNLGSAYVIPQFFQEAFQQEEPIENYLPQLNDDDSSAVAANIPKP HIPYFMKPHVESEKLQDKIKVDDLNATAWDLYRLANYSTPDYGHPTRVIGSKGHNKTMEY ILNVFDDMQDYYDVSLQEFEALSGKIISFNLSDAETGKSFANTTAFALSPPVDGFVGKLV EIPNLGCEEKDYASVVPPRHNEKQIALIERGKCPFGDKSNLAGKFGFTAVVIYDNEPKSK EGLHGTLGEPTKHTVATVGVPYKVGKKLIANIALNIDYSLYFAMDSYVEFIKTQNIIADT KHGDPDNIVALGAHSDSVEEGPGINDDGSGTISLLNVAKQLTHFKINNKVRFAWWAAEEE GLLGSNFYAYNLTKEENSKIRVFMDYDMMASPNYEYEIYDANNKENPKGSEELKNLYVDY YKAHHLNYTLVPFDGRSDYVGFINNGIPAGGIATGAEKNNVNNGKVLDRCYHQLCDDVSN LSWDAFITNTKLIAHSVATYADSFEGFPKRETQKHKEVDILNAQQPQFKYRADFLII
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| YBR286W | 239 S | NEPKSKEGL | 0.995 | unsp | YBR286W | 317 S | AHSDSVEEG | 0.998 | unsp |