

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
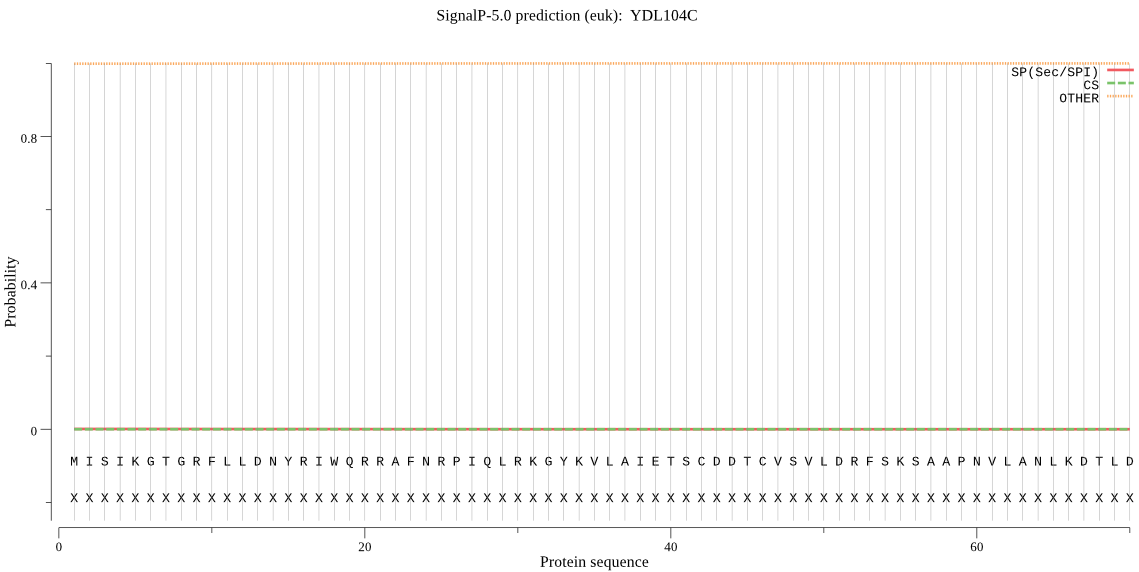
| YDL104C | OTHER | 0.924928 | 0.002134 | 0.072938 |
MISIKGTGRFLLDNYRIWQRRAFNRPIQLRKGYKVLAIETSCDDTCVSVLDRFSKSAAPN VLANLKDTLDSIDEGGIIPTKAHIHHQARIGPLTERALIESNAREGIDLICVTRGPGMPG SLSGGLDFAKGLAVAWNKPLIGVHHMLGHLLIPRMGTNGKVPQFPFVSLLVSGGHTTFVL SRAIDDHEILCDTIDIAVGDSLDKCGRELGFKGTMIAREMEKFINQDINDQDFALKLEMP SPLKNSASKRNMLSFSFSAFITALRTNLTKLGKTEIQELPEREIRSIAYQVQESVFDHII NKLKHVLKSQPEKFKNVREFVCSGGVSSNQRLRTKLETELGTLNSTSFFNFYYPPMDLCS DNSIMIGWAGIEIWESLRLVSDLDICPIRQWPLNDLLSVDGWRTDQL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| YDL104C | 294 S | QVQESVFDH | 0.992 | unsp |