

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
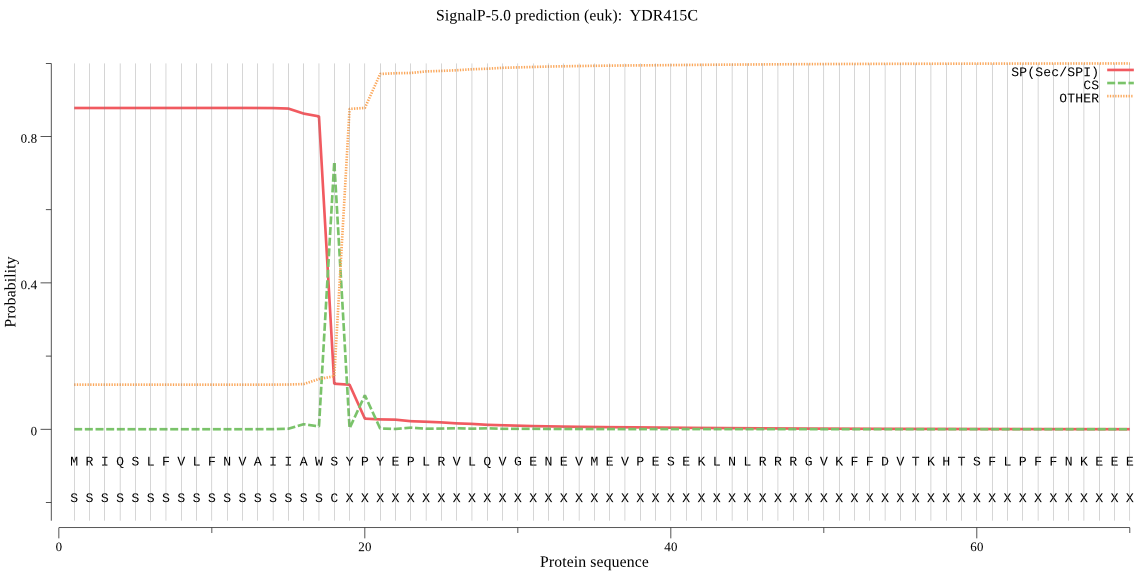
| YDR415C | SP | 0.010081 | 0.989426 | 0.000492 | CS pos: 18-19. AWS-YP. Pr: 0.7553 |
MRIQSLFVLFNVAIIAWSYPYEPLRVLQVGENEVMEVPESEKLNLRRRGVKFFDVTKHTS FLPFFNKEEEPTVPTYNYPPEISNKEVVDDSIKNIDKGSMHKNLAKFTSFYTRYYKSDHG FESAEWLAATIANITKDIPQDTLTIEHFDHKEWKQYSIIVRVTGSTTPEDIIIIGSHQDS INLLLPSIMAAPGADDNGSGTVTNMEALRLYTENFLKRGFRPNNTVEFHFYSAEEGGLLG SLDVFTAYAKQKKHVRAMLQQDMTGYVSDPEDEHVGIVTDYTTPALTDFIKLIINSYLSI PYRDTQCGYACSDHGSATRNGFPGSFVIESEFKKTNKYIHSTMDTLDRLSLAHMAEHTKI VLGVIIELGSWSAW
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| YDR415C | 91 S | VVDDSIKNI | 0.995 | unsp |