

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YFL060C | OTHER | 0.858271 | 0.140912 | 0.000817 |
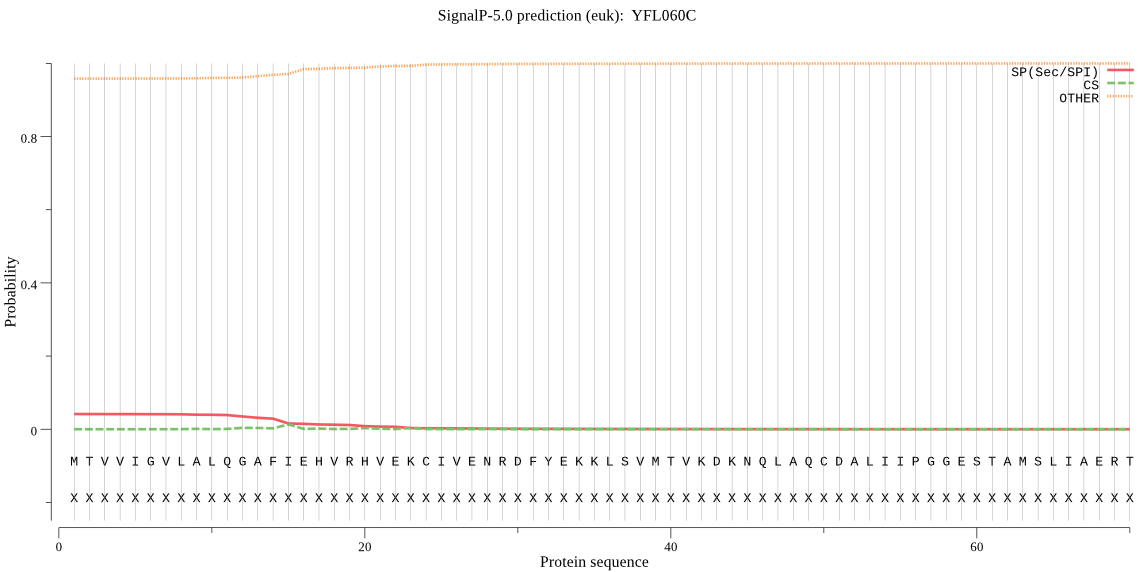
MTVVIGVLALQGAFIEHVRHVEKCIVENRDFYEKKLSVMTVKDKNQLAQCDALIIPGGES TAMSLIAERTGFYDDLYAFVHNPSKVTWGTCAGLIYISQQLSNEAKLVKTLNLLKVKVKR NAFGRQAQSSTRICDFSNFIPHCNDFPATFIRAPVIEEVLDPEHVQVLYKLDGKDNGGQE LIVAAKQKNNILATSFHPELAENDIRFHDWFIREFVLKNYSK