

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YFR018C | SP | 0.059078 | 0.940491 | 0.000431 | CS pos: 28-29. VTG-WE. Pr: 0.9792 |
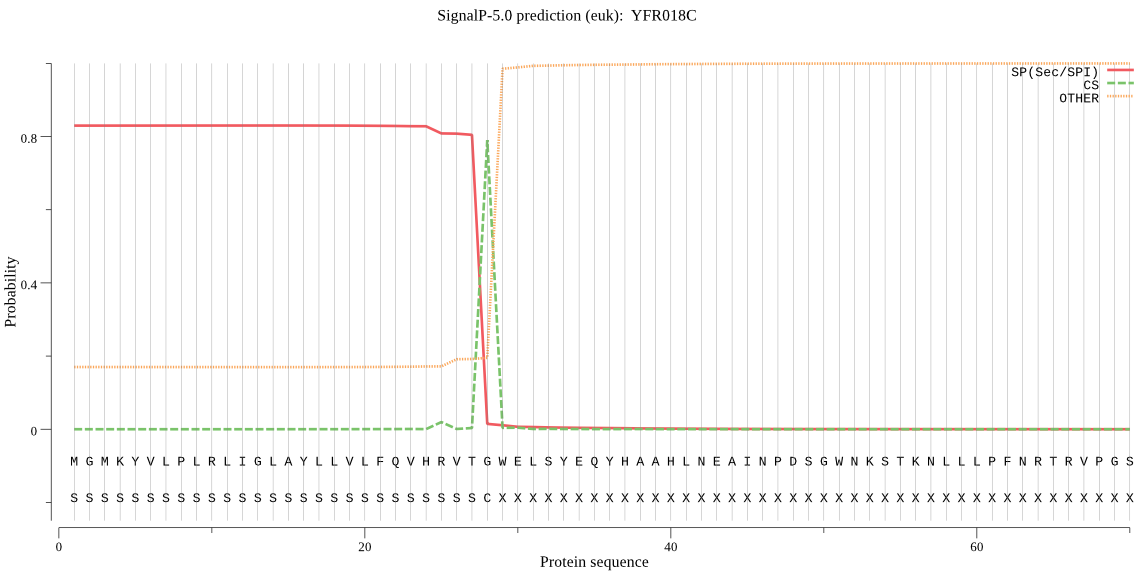
MGMKYVLPLRLIGLAYLLVLFQVHRVTGWELSYEQYHAAHLNEAINPDSGWNKSTKNLLL PFNRTRVPGSEGSREIQRFIIEHFNNTLAGEWAVETQAFEENGYRFNNLVMTLQNNASEY LVLAAHYDTKIAPTGMVGAIDSAASCAALLYTAQFLTHIACHERTKEYNDLESNTVVSNS TLGVKIVFFDGEEAIEEWGPEDSIYGARRLAAQWLADGTMTRIRLLFLLDLLGSGEEEPL VPSYYAETHQEYQLLNRIEDDLLFRRGDEINGESALAAEVARQRKHLDPTDYRFLGLGHS VIGDDHTPFLAAGVPVLHAIPLPFPSTWHTVDDDFRHLDAAETRHWALLVCEFVVQSLRS RNQ