

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
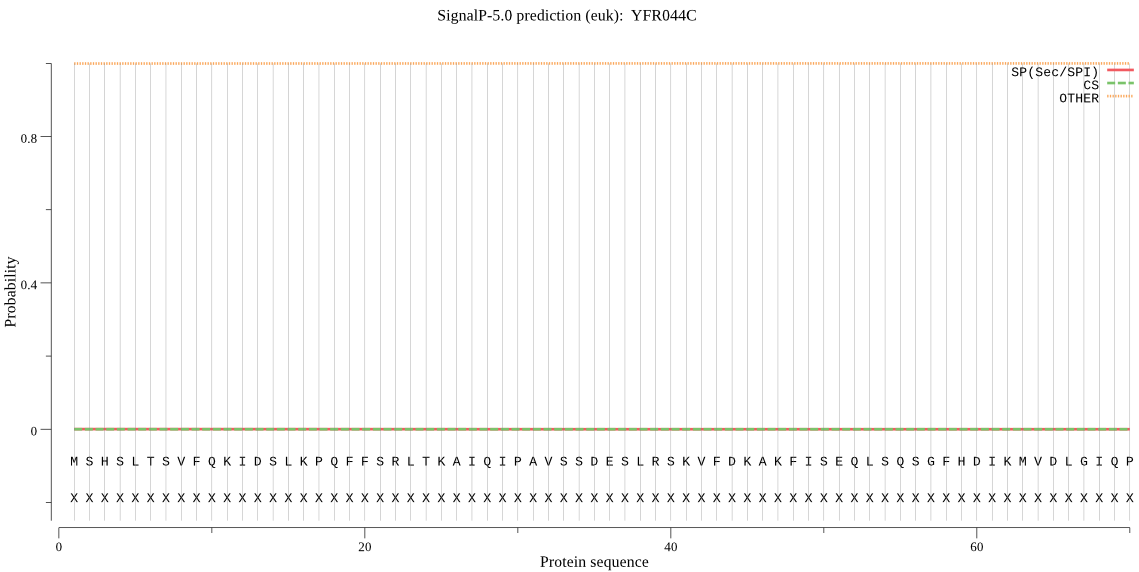
| YFR044C | OTHER | 0.949658 | 0.001357 | 0.048984 |
MSHSLTSVFQKIDSLKPQFFSRLTKAIQIPAVSSDESLRSKVFDKAKFISEQLSQSGFHD IKMVDLGIQPPPISTPNLSLPPVILSRFGSDPSKKTVLVYGHYDVQPAQLEDGWDTEPFK LVIDEAKGIMKGRGVTDDTGPLLSWINVVDAFKASGQEFPVNLVTCFEGMEESGSLKLDE LIKKEANGYFKGVDAVCISDNYWLGTKKPVLTYGLRGCNYYQTIIEGPSADLHSGIFGGV VAEPMIDLMQVLGSLVDSKGKILIDGIDEMVAPLTEKEKALYKDIEFSVEELNAATGSKT SLYDKKEDILMHRWRYPSLSIHGVEGAFSAQGAKTVIPAKVFGKFSIRTVPDMDSEKLTS LVQKHCDAKFKSLNSPNKCRTELIHDGAYWVSDPFNAQFTAAKKATKLVYGVDPDFTREG GSIPITLTFQDALNTSVLLLPMGRGDDGAHSINEKLDISNFVGGMKTMAAYLQYYSESPE N
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| YFR044C | 301 S | GSKTSLYDK | 0.996 | unsp |