

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YGL203C | SP | 0.028332 | 0.964302 | 0.007367 | CS pos: 22-23. SVS-LP. Pr: 0.7657 |
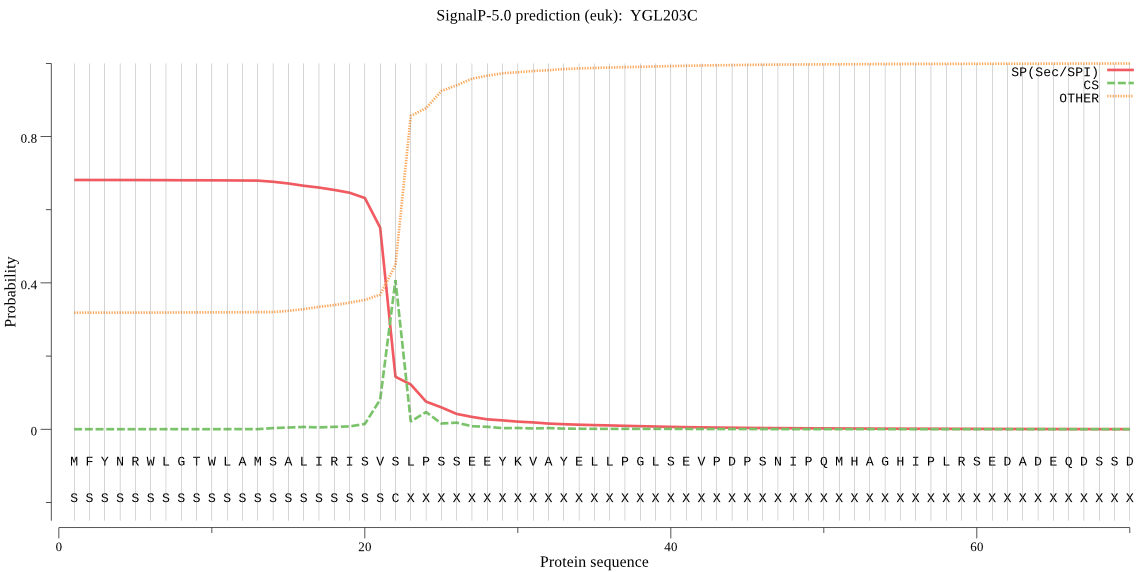
MFYNRWLGTWLAMSALIRISVSLPSSEEYKVAYELLPGLSEVPDPSNIPQMHAGHIPLRS EDADEQDSSDLEYFFWKFTNNDSNGNVDRPLIIWLNGGPGCSSMDGALVESGPFRVNSDG KLYLNEGSWISKGDLLFIDQPTGTGFSVEQNKDEGKIDKNKFDEDLEDVTKHFMDFLENY FKIFPEDLTRKIILSGESYAGQYIPFFANAILNHNKFSKIDGDTYDLKALLIGNGWIDPN TQSLSYLPFAMEKKLIDESNPNFKHLTNAHENCQNLINSASTDEAAHFSYQECENILNLL LSYTRESSQKGTADCLNMYNFNLKDSYPSCGMNWPKDISFVSKFFSTPGVIDSLHLDSDK IDHWKECTNSVGTKLSNPISKPSIHLLPGLLESGIEIVLFNGDKDLICNNKGVLDTIDNL KWGGIKGFSDDAVSFDWIHKSKSTDDSEEFSGYVKYDRNLTFVSVYNASHMVPFDKSLVS RGIVDIYSNDVMIIDNNGKNVMITTDDDSDQDATTESGDKPKENLEEEEQEAQNEEGKEK EGNKDKDGDDDNDNDDDDEDDHNSEGDDDDDDDDDEDDNNEKQSNQGLEDSRHKSSEYEQ EEEEVEEFAEEISMYKHKAVVVTIVTFLIVVLGVYAYDRRVRRKARHTILVDPNNRQHDS PNKTVSWADDLESGLGAEDDLEQDEQLEGGAPISSTSNKAGSKLKTKKKKKYTSLPNTEI DESFEMTDF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YGL203C | 308 S | TRESSQKGT | 0.997 | unsp | YGL203C | 308 S | TRESSQKGT | 0.997 | unsp | YGL203C | 308 S | TRESSQKGT | 0.997 | unsp | YGL203C | 443 S | HKSKSTDDS | 0.998 | unsp | YGL203C | 447 S | STDDSEEFS | 0.994 | unsp | YGL203C | 509 S | TDDDSDQDA | 0.997 | unsp | YGL203C | 564 S | DDHNSEGDD | 0.994 | unsp | YGL203C | 596 S | RHKSSEYEQ | 0.997 | unsp | YGL203C | 666 S | NKTVSWADD | 0.993 | unsp | YGL203C | 714 S | KKYTSLPNT | 0.991 | unsp | YGL203C | 723 S | EIDESFEMT | 0.996 | unsp | YGL203C | 25 S | VSLPSSEEY | 0.996 | unsp | YGL203C | 218 S | HNKFSKIDG | 0.99 | unsp |