

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YGR101W | mTP | 0.070484 | 0.000087 | 0.929429 | CS pos: 39-40. RSQ-ST. Pr: 0.3568 |
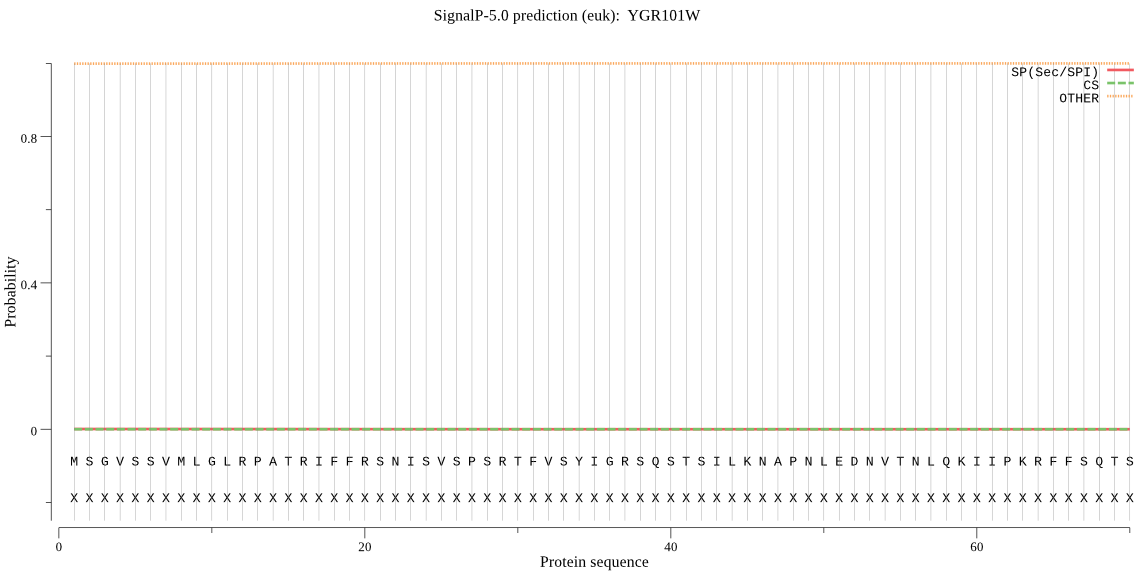
MSGVSSVMLGLRPATRIFFRSNISVSPSRTFVSYIGRSQSTSILKNAPNLEDNVTNLQKI IPKRFFSQTSILKSRWKPIFNEETTNRYVRLNRFQQYQQQRSGGNPLGSMTILGLSLMAG IYFGSPYLFEHVPPFTYFKTHPKNLVYALLGINVAVFGLWQLPKCWRFLQKYMLLQKDYV TSKISIIGSAFSHQEFWHLGMNMLALWSFGTSLATMLGASNFFSLYMNSAIAGSLFSLWY PKLARLAIVGPSLGASGALFGVLGCFSYLFPHAKILLFVFPVPGGAWVAFLASVAWNAAG CALRWGSFDYAAHLGGSMMGVLYGWYISKAVEKQRQRRLQAAGRWF
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| YGR101W | 26 S | NISVSPSRT | 0.997 | unsp |