

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YHR132C | SP | 0.002246 | 0.997721 | 0.000033 | CS pos: 24-25. VQG-LQ. Pr: 0.6399 |
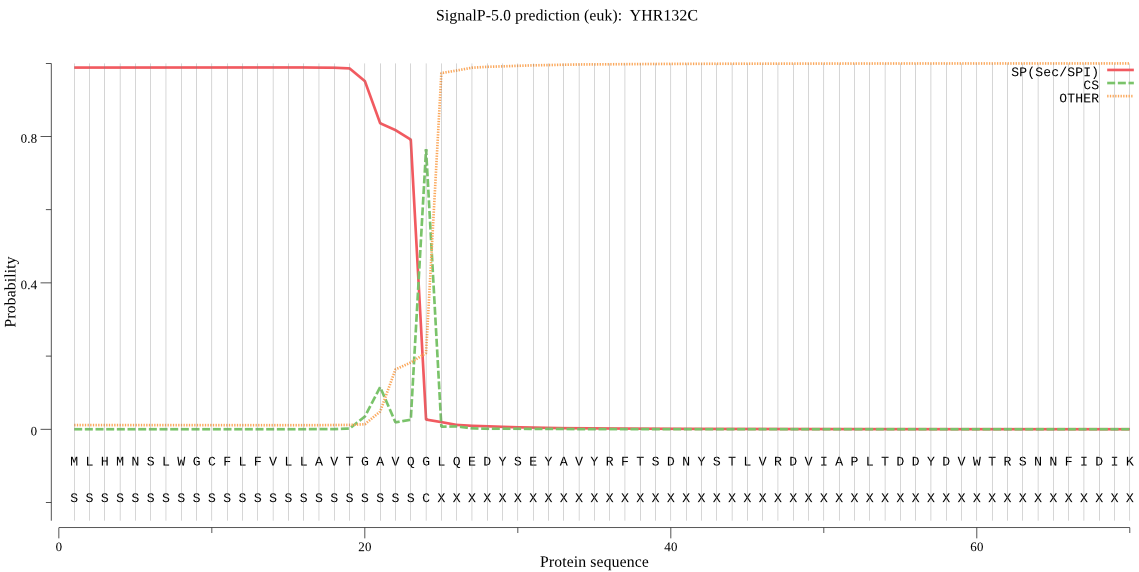
MLHMNSLWGCFLFVLLAVTGAVQGLQEDYSEYAVYRFTSDNYSTLVRDVIAPLTDDYDVW TRSNNFIDIKLPKEIGEQINDGQVIIDNMNELIQNTLPTSQMMAREQAVFENDYDFFFNE YRDLDTIYMWLDLLERSFPSLVAVEHLGRTFEGRELKALHISGNKPESNPEKKTIVITGG IHAREWISVSTVCWALYQLLNRYGSSKKETKYLDDLDFLVIPVFNPDGYAYTWSHDRLWR KNRQRTHVPQCLGIDIDHSFGFQWEKAHTHACSEEYSGETPFEAWEASAWYKYINETKGD YKIYGYIDMHSYSQEILYPYAYSCDALPRDLENLLELSYGLSKAIRSKSGRNYDVISACK DRGSDIFPGLGAGSALDFMYHHRAHWAFQLKLRDTGNHGFLLPPENIKPVGKETYAALKY FCDFLLDPEI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YHR132C | 349 S | IRSKSGRNY | 0.997 | unsp | YHR132C | 349 S | IRSKSGRNY | 0.997 | unsp | YHR132C | 349 S | IRSKSGRNY | 0.997 | unsp | YHR132C | 205 S | NRYGSSKKE | 0.991 | unsp | YHR132C | 206 S | RYGSSKKET | 0.993 | unsp |