

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
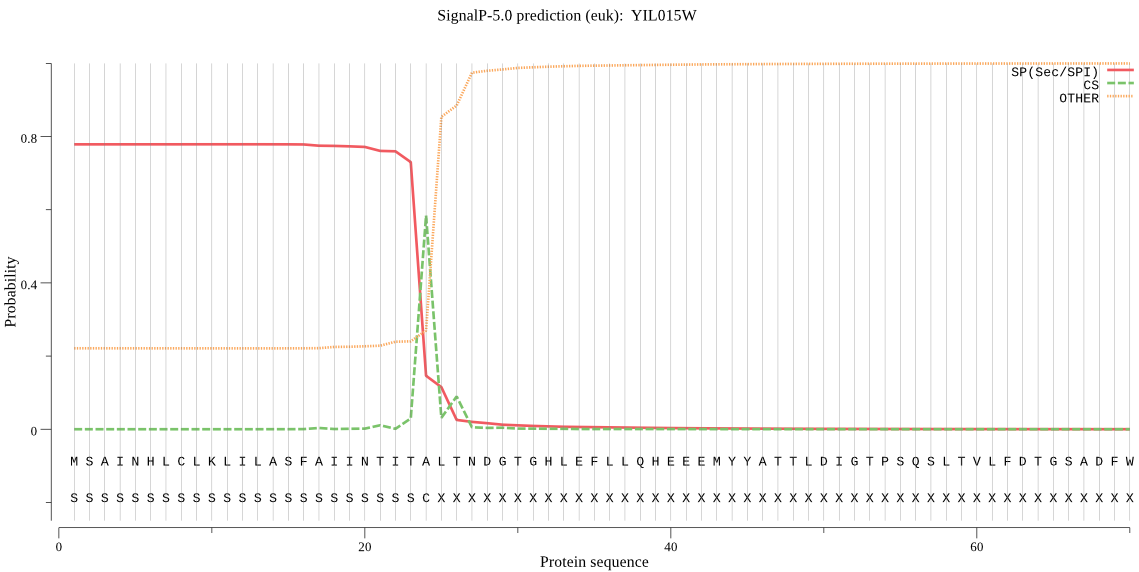
| YIL015W | SP | 0.009801 | 0.990147 | 0.000052 | CS pos: 24-25. ITA-LT. Pr: 0.8363 |
MSAINHLCLKLILASFAIINTITALTNDGTGHLEFLLQHEEEMYYATTLDIGTPSQSLTV LFDTGSADFWVMDSSNPFCLPNSNTSSYSNATYNGEEVKPSIDCRSMSTYNEHRSSTYQY LENGRFYITYADGTFADGSWGTETVSINGIDIPNIQFGVAKYATTPVSGVLGIGFPRRES VKGYEGAPNEYYPNFPQILKSEKIIDVVAYSLFLNSPDSGTGSIVFGAIDESKFSGDLFT FPMVNEYPTIVDAPATLAMTIQGLGAQNKSSCEHETFTTTKYPVLLDSGTSLLNAPKVIA DKMASFVNASYSEEEGIYILDCPVSVGDVEYNFDFGDLQISVPLSSLILSPETEGSYCGF AVQPTNDSMVLGDVFLSSAYVVFDLDNYKISLAQANWNASEVSKKLVNIQTDGSISGAKI ATAEPWSTNEPFTVTSDIYSSTGCKSRPFLQSSTASSLIAETNVQSRNCSTKMPGTRSTT VLSKPTQNSAMHQSTGAVTQTSNETKLELSSTMANSGSVSLPTSNSIDKEFEHSKSQTTS DPSVAEHSTFNQTFVHETKYRPTHKTVITETVTKYSTVLINVCKPTY
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| YIL015W | T | 428 | 0.501 | 0.254 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| YIL015W | T | 428 | 0.501 | 0.254 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YIL015W | 312 S | NASYSEEEG | 0.995 | unsp | YIL015W | 312 S | NASYSEEEG | 0.995 | unsp | YIL015W | 312 S | NASYSEEEG | 0.995 | unsp | YIL015W | 543 S | TSDPSVAEH | 0.996 | unsp | YIL015W | 108 S | CRSMSTYNE | 0.991 | unsp | YIL015W | 180 S | PRRESVKGY | 0.998 | unsp |