

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
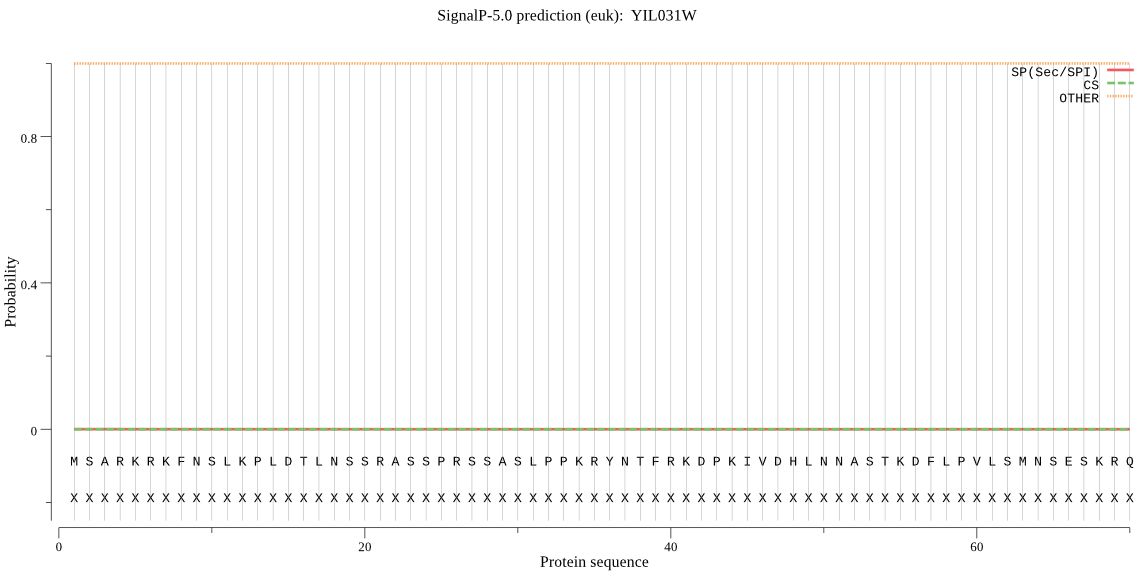
| YIL031W | OTHER | 0.998844 | 0.000017 | 0.001139 |
MSARKRKFNSLKPLDTLNSSRASSPRSSASLPPKRYNTFRKDPKIVDHLNNASTKDFLPV LSMNSESKRQIELSDNDVDNNDEGEGVNSGCSDQDFEPLQSSPLKRHSSLKSTSNGLLFQ MSNNLGNGSPEPAVASTSPNGSIISTKLNLNGQFSCVDSKTLRIYRHKAPCIMTFVSDHN HPKFSLYFQQSVIYNSQVNLLDDVELIILDKKNSFMAIILKDLKKVKMILDVNNSSININ TNILIWSTASSASNKKIKSIKRFLLMSYSSSIKVEILDHKEQILERLKHLIHPISSSSPS LNMERAINSTKNAFDSLRLKKTKLSTNDDESPQIHTHFLSNKPHGLQSLTKRTRIASLGK KEHSISVPKSNISPSDFYNTNGTETLQSHAVSQLRRSNRFKDVSDPANSNSNSEFDDATT EFETPELFKPSLCYKFNDGSSYTITNQDFKCLFNKDWVNDSILDFFTKFYIESSIEKSII KREQVHLMSSFFYTKLISNPADYYSNVKKWVNNTDLFSKKYVVIPINISYHWFSCIITNL DAILDFHQNKDKNDAINSDEISINNPLVNILTFDSLRQTHSREIDPIKEFLISYALDKYS IQLDKTQIKMKTCPVPQQPNMSDCGVHVILNIRKFFENPVETIDVWKNSKIKSKHFTAKM INKYFDKNERNSARKNLRHTLKLLQLNYISYLKKENLYEEVMQMEEKKSTNINNNENYDD DDEEIQIIENIDQSSKDNNAQLTSEPPCSRSSSISTTEREPTELHNSVVRQPTGEIITDN EDPVRAASPETASVSPPIRHNILKSSSPFISESANETEQEEFTSPYFGRPSLKTRAKQFE GVSSPIKNDQALSSTHDIMMPSPKPKRIYPSKKIPQLSSHVQSLSTDSMERQSSPNNTNI VISDTEQDSRLGVNSESKNTSGIVNRDDSDVNLIGSSLPNVAEKNHDNTQESNGNNDSLG KILQNVDKELNEKLVDIDDVAFSSPTRGIPRTSATSKGSNAQLLSNYGDENNQSQDSVWD EGRDNPILLEDEDP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YIL031W | 109 S | KRHSSLKST | 0.994 | unsp | YIL031W | 109 S | KRHSSLKST | 0.994 | unsp | YIL031W | 109 S | KRHSSLKST | 0.994 | unsp | YIL031W | 253 S | ASSASNKKI | 0.991 | unsp | YIL031W | 325 S | KTKLSTNDD | 0.996 | unsp | YIL031W | 373 S | KSNISPSDF | 0.997 | unsp | YIL031W | 397 S | QLRRSNRFK | 0.995 | unsp | YIL031W | 672 S | NERNSARKN | 0.996 | unsp | YIL031W | 734 S | NIDQSSKDN | 0.994 | unsp | YIL031W | 752 S | CSRSSSIST | 0.993 | unsp | YIL031W | 755 S | SSSISTTER | 0.996 | unsp | YIL031W | 788 S | VRAASPETA | 0.997 | unsp | YIL031W | 888 S | LSTDSMERQ | 0.995 | unsp | YIL031W | 894 S | ERQSSPNNT | 0.995 | unsp | YIL031W | 903 S | NIVISDTEQ | 0.995 | unsp | YIL031W | 996 S | TSATSKGSN | 0.992 | unsp | YIL031W | 1017 S | QSQDSVWDE | 0.99 | unsp | YIL031W | 23 S | SSRASSPRS | 0.991 | unsp | YIL031W | 24 S | SRASSPRSS | 0.998 | unsp |