

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YIR022W | SP | 0.250294 | 0.733800 | 0.015906 | CS pos: 30-31. AIA-TN. Pr: 0.4858 |
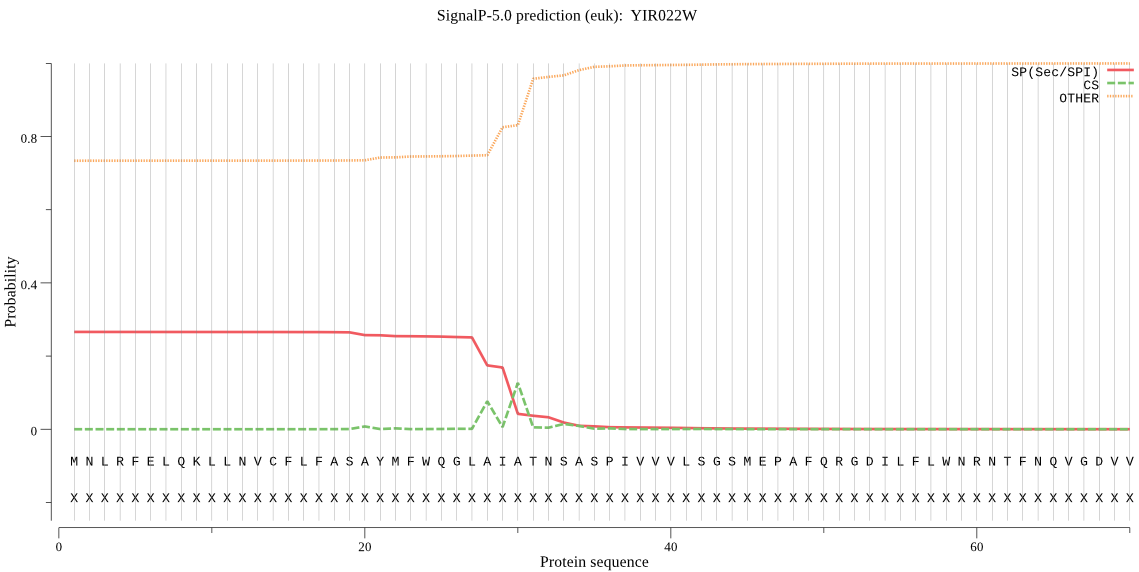
MNLRFELQKLLNVCFLFASAYMFWQGLAIATNSASPIVVVLSGSMEPAFQRGDILFLWNR NTFNQVGDVVVYEVEGKQIPIVHRVLRQHNNHADKQFLLTKGDNNAGNDISLYANKKIYL NKSKEIVGTVKGYFPQLGYITIWISENKYAKFALLGMLGLSALLGGE