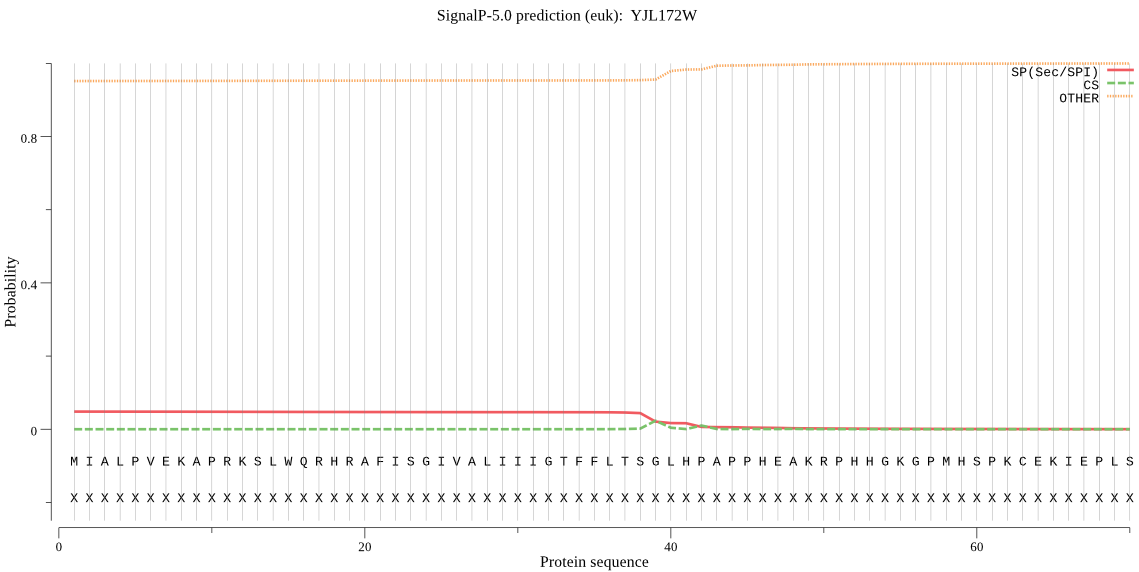
Fasta :-
MIALPVEKAPRKSLWQRHRAFISGIVALIIIGTFFLTSGLHPAPPHEAKRPHHGKGPMHS
PKCEKIEPLSPSFKHSVDTILHDPAFRNSSIEKLSNAVRIPTVVQDKNPNPADDPDFYKH
FYELHDYFEKTFPNIHKHLKLEKVNELGLLYTWEGSDPDLKPLLLMAHQDVVPVNNETLS
SWKFPPFSGHYDPETDFVWGRGSNDCKNLLIAEFEAIEQLLIDGFKPNRTIVMSLGFDEE
ASGTLGAASLASFLHERYGDDGIYSIIDEGEGIMEVDKDVFVATPINAEKGYVDFEVSIL
GHGGHSSVPPDHTTIGIASELITEFEANPFDYEFEFDNPIYGLLTCAAEHSKSLSKDVKK
TILGAPFCPRRKDKLVEYISNQSHLRSLIRTTQAVDIINGGVKANALPETTRFLINHRIN
LHSSVAEVFERNIEYAKKIAEKYGYGLSKNGDDYIIPETELGHIDITLLRELEPAPLSPS
SGPVWDILAGTIQDVFENGVLQNNEEFYVTTGLFSGNTDTKYYWNLSKNIYRFVGSIIDI
DLLKTLHSVNEHVDVPGHLSAIAFVYEYIVNVNEYA