

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YKL134C | mTP | 0.101903 | 0.000416 | 0.897681 | CS pos: 28-29. RFF-AT. Pr: 0.7154 |
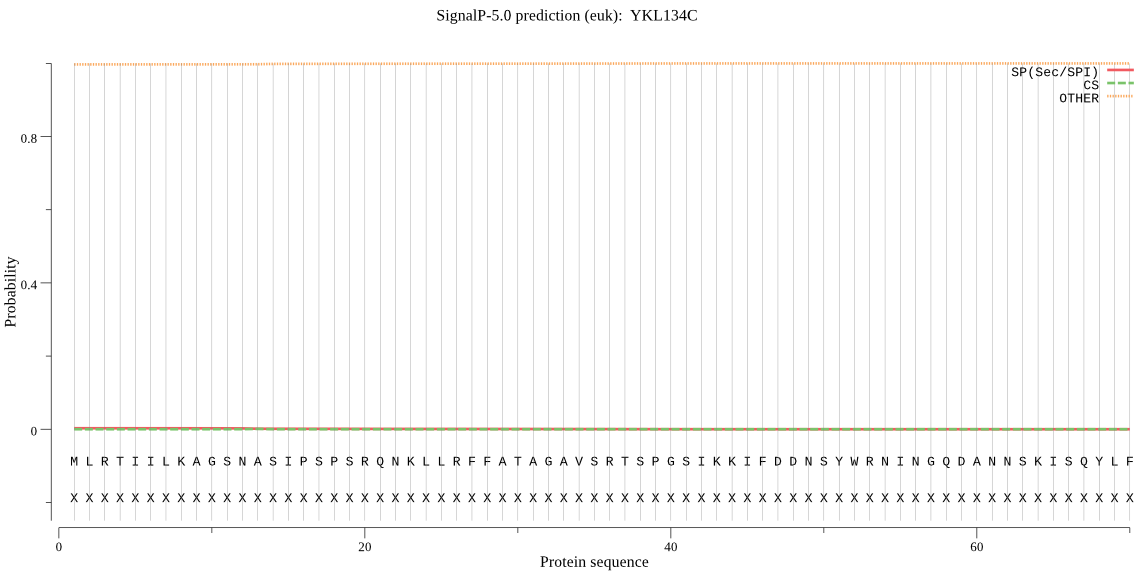
MLRTIILKAGSNASIPSPSRQNKLLRFFATAGAVSRTSPGSIKKIFDDNSYWRNINGQDA NNSKISQYLFKKNKTGLFKNPYLTSPDGLRKFSQVSLQQAQELLDKMRNDFSESGKLTYI MNLDRLSDTLCRVIDLCEFIRSTHPDDAFVRAAQDCHEQMFEFMNVLNTDVSLCNILKSV LNNPEVSSKLSAEELKVGKILLDDFEKSGIYMNPDVREKFIQLSQEISLVGQEFINHTDY PGSNSVKIPCKDLDNSKVSTFLLKQLNKDVKGQNYKVPTFGYAAYALLKSCENEMVRKKL WTALHSCSDKQVKRLSHLIKLRAILANLMHKTSYAEYQLEGKMAKNPKDVQDFILTLMNN TIEKTANELKFIAELKAKDLKKPLTTNTDEILKLVRPWDRDYYTGKYFQLNPSNSPNAKE ISYYFTLGNVIQGLSDLFQQIYGIRLEPAITDEGETWSPDVRRLNVISEEEGIIGIIYCD LFERNGKTSNPAHFTVCCSRQIYPSETDFSTIQVGENPDGTYFQLPVISLVCNFSPILIA SKKSLCFLQLSEVETLFHEMGHAMHSMLGRTHMQNISGTRCATDFVELPSILMEHFAKDI RILTKIGKHYGTGETIQADMLQRFMKSTNFLQNCETYSQAKMAMLDQSFHDEKIISDIDN FDVVENYQALERRLKVLVDDQSNWCGRFGHLFGYGATYYSYLFDRTIASKIWYALFEDDP YSRKNGDKFKKHLLKWGGLKDPWKCIADVLECPMLEKGGSDAMEFIAQSHKS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YKL134C | 191 S | SSKLSAEEL | 0.996 | unsp | YKL134C | 191 S | SSKLSAEEL | 0.996 | unsp | YKL134C | 191 S | SSKLSAEEL | 0.996 | unsp | YKL134C | 468 S | LNVISEEEG | 0.992 | unsp | YKL134C | 505 S | QIYPSETDF | 0.995 | unsp | YKL134C | 656 S | EKIISDIDN | 0.991 | unsp | YKL134C | 722 S | DDPYSRKNG | 0.996 | unsp | YKL134C | 38 S | VSRTSPGSI | 0.997 | unsp | YKL134C | 41 S | TSPGSIKKI | 0.995 | unsp |