

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YKR038C | OTHER | 0.999371 | 0.000515 | 0.000113 |
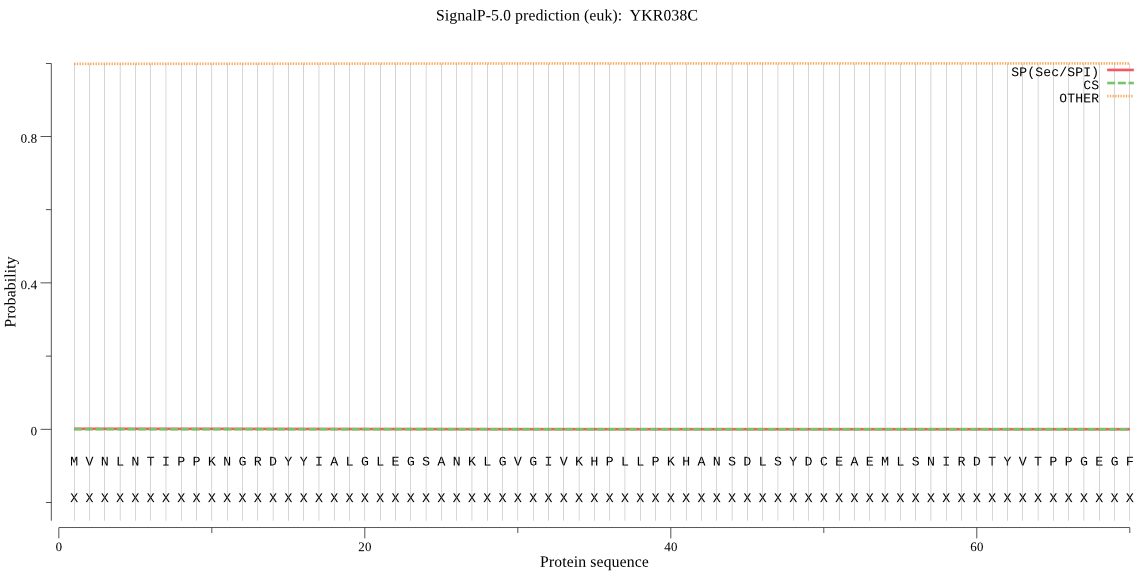
MVNLNTIPPKNGRDYYIALGLEGSANKLGVGIVKHPLLPKHANSDLSYDCEAEMLSNIRD TYVTPPGEGFLPRDTARHHRNWCIRLIKQALAEADIKSPTLDIDVICFTKGPGMGAPLHS VVIAARTCSLLWDVPLVGVNHCIGHIEMGREITKAQNPVVLYVSGGNTQVIAYSEKRYRI FGETLDIAIGNCLDRFARTLKIPNEPSPGYNIEQLAKKAPHKENLVELPYTVKGMDLSMS GILASIDLLAKDLFKGNKKNKILFDKTTGEQKVTVEDLCYSLQENLFAMLVEITERAMAH VNSNQVLIVGGVGCNVRLQEMMAQMCKDRANGQVHATDNRFCIDNGVMIAQAGLLEYRMG GIVKDFSETVVTQKFRTDEVYAAWRD