

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YLL029W | OTHER | 0.999953 | 0.000023 | 0.000024 |
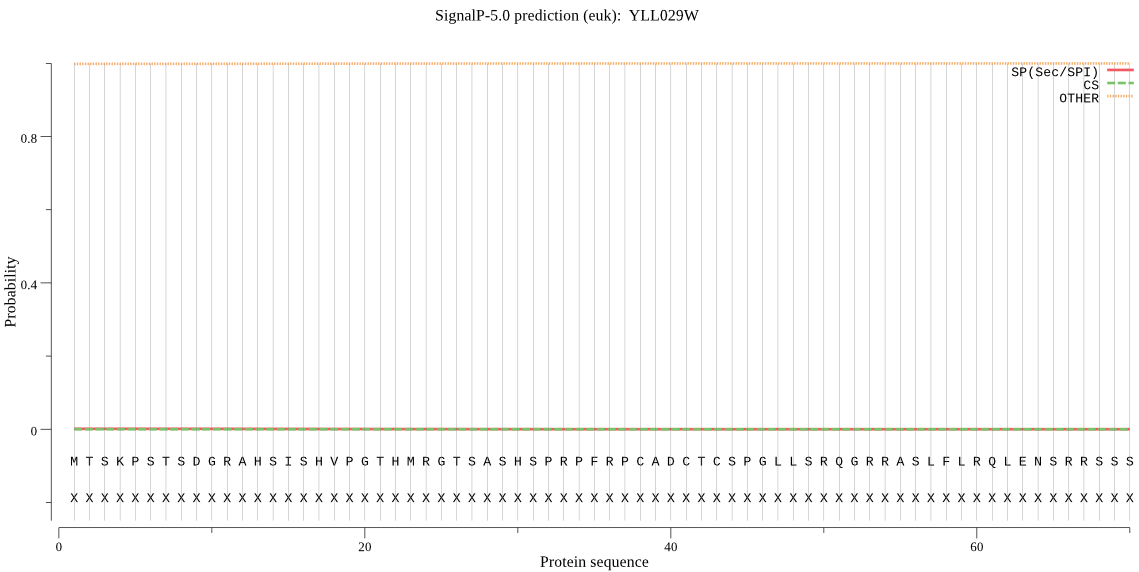
MTSKPSTSDGRAHSISHVPGTHMRGTSASHSPRPFRPCADCTCSPGLLSRQGRRASLFLR QLENSRRSSSMLLNELKGAGGGSSAGNGSVYSCDSLCAVNREVNTTDRLLKLRQEMKKHD LCCYIVPSCDEHQSEYVSLRDQRRAFISGFSGSAGVACITRDLLNFNDDHPDGKSILSTD GRYFNQARQELDYNWTLLRQNEDPITWQEWCVREALEMAKGLGNKEGMVLKIGIDPKLIT FNDYVSFRKMIDTKYDAKGKVELVPVEENLVDSIWPDFETLPERPCNDLLLLKYEFHGEE FKDKKEKLLKKLNDKASSATTGRNTFIVVALDEICWLLNLRGSDIDYNPVFFSYVAINED ETILFTNNPFNDDISEYFKINGIEVRPYEQIWEHLTKITSQASSAEHEFLIPDSASWQMV RCLNTSTNANGAIAKKMTAQNFAIIHSPIDVLKSIKNDIEIKNAHKAQVKDAVCLVQYFA WLEQQLVGREALIDEYRAAEKLTEIRKTQRNFMGNSFETISSTGSNAAIIHYSPPVENSS MIDPTKIYLCDSGSQFLEGTTDITRTIHLTKPTKEEMDNYTLVLKGGLALERLIFPENTP GFNIDAIARQFLWSRGLDYKHGTGHGIGSFLNVHEGPMGVGFRPHLMNFPLRAGNIISNE PGYYKDGEYGIRIESDMLIKKATEKGNFLKFENMTVVPYCRKLINTKLLNEEEKTQINEY HARVWRTIVHFLQPQSISYKWLKRETSPL
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YLL029W | T | 2 | 0.613 | 0.640 | YLL029W | T | 7 | 0.568 | 0.050 | YLL029W | S | 3 | 0.505 | 0.068 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YLL029W | T | 2 | 0.613 | 0.640 | YLL029W | T | 7 | 0.568 | 0.050 | YLL029W | S | 3 | 0.505 | 0.068 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YLL029W | 68 S | NSRRSSSML | 0.994 | unsp | YLL029W | 68 S | NSRRSSSML | 0.994 | unsp | YLL029W | 68 S | NSRRSSSML | 0.994 | unsp | YLL029W | 138 S | SEYVSLRDQ | 0.997 | unsp | YLL029W | 403 S | TSQASSAEH | 0.993 | unsp | YLL029W | 404 S | SQASSAEHE | 0.992 | unsp | YLL029W | 454 S | DVLKSIKND | 0.991 | unsp | YLL029W | 540 S | VENSSMIDP | 0.994 | unsp | YLL029W | 6 S | TSKPSTSDG | 0.998 | unsp | YLL029W | 31 S | SASHSPRPF | 0.997 | unsp |