

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YLR120C | SP | 0.131772 | 0.844635 | 0.023593 | CS pos: 21-22. VLG-KI. Pr: 0.8567 |
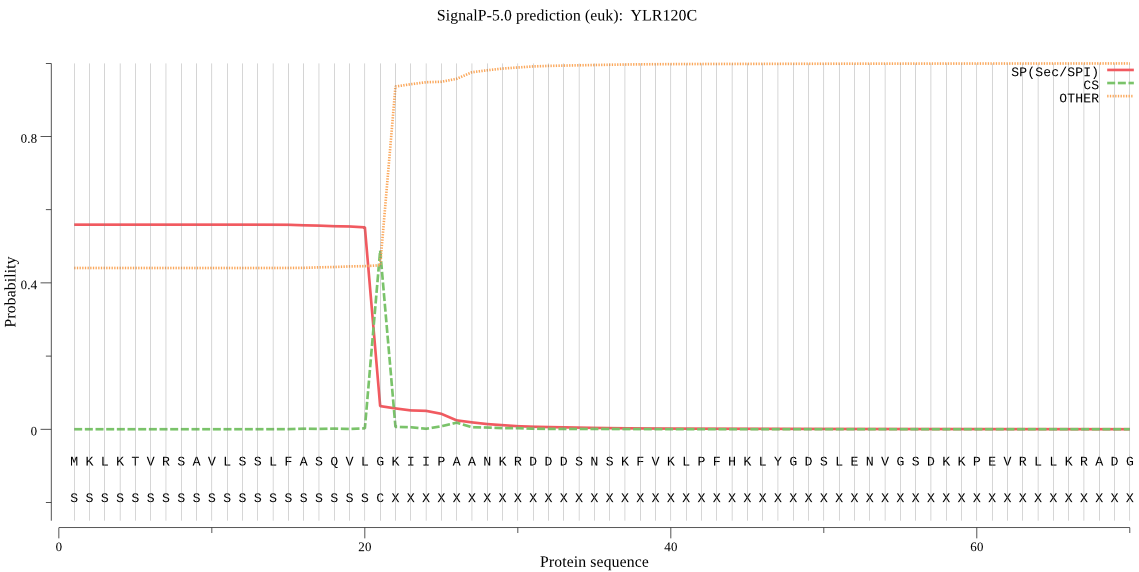
MKLKTVRSAVLSSLFASQVLGKIIPAANKRDDDSNSKFVKLPFHKLYGDSLENVGSDKKP EVRLLKRADGYEEIIITNQQSFYSVDLEVGTPPQNVTVLVDTGSSDLWIMGSDNPYCSSN SMGSSRRRVIDKRDDSSSGGSLINDINPFGWLTGTGSAIGPTATGLGGGSGTATQSVPAS EATMDCQQYGTFSTSGSSTFRSNNTYFSISYGDGTFASGTFGTDVLDLSDLNVTGLSFAV ANETNSTMGVLGIGLPELEVTYSGSTASHSGKAYKYDNFPIVLKNSGAIKSNTYSLYLND SDAMHGTILFGAVDHSKYTGTLYTIPIVNTLSASGFSSPIQFDVTINGIGISDSGSSNKT LTTTKIPALLDSGTTLTYLPQTVVSMIATELGAQYSSRIGYYVLDCPSDDSMEIVFDFGG FHINAPLSSFILSTGTTCLLGIIPTSDDTGTILGDSFLTNAYVVYDLENLEISMAQARYN TTSENIEIITSSVPSAVKAPGYTNTWSTSASIVTGGNIFTVNSSQTASFSGNLTTSTASA TSTSSKRNVGDHIVPSLPLTLISLLFAFI
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| YLR120C | T | 172 | 0.517 | 0.026 | YLR120C | T | 164 | 0.513 | 0.231 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| YLR120C | T | 172 | 0.517 | 0.026 | YLR120C | T | 164 | 0.513 | 0.231 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YLR120C | 270 S | TASHSGKAY | 0.995 | unsp | YLR120C | 270 S | TASHSGKAY | 0.995 | unsp | YLR120C | 270 S | TASHSGKAY | 0.995 | unsp | YLR120C | 542 S | ASATSTSSK | 0.991 | unsp | YLR120C | 544 S | ATSTSSKRN | 0.995 | unsp | YLR120C | 136 S | KRDDSSSGG | 0.996 | unsp | YLR120C | 210 S | YFSISYGDG | 0.996 | unsp |