

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
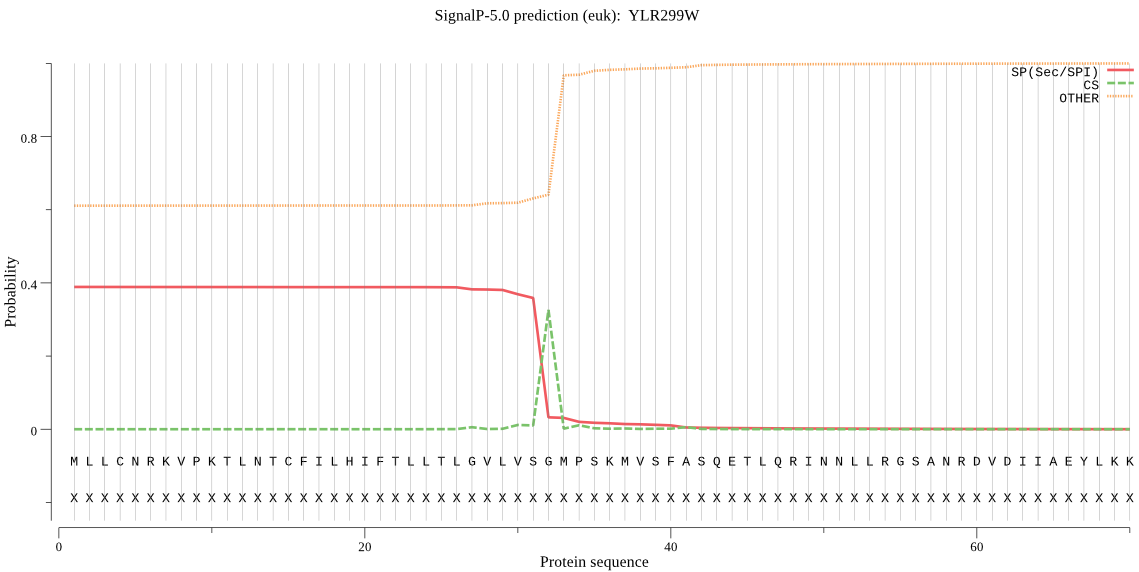
| YLR299W | SP | 0.094684 | 0.897575 | 0.007741 | CS pos: 32-33. VSG-MP. Pr: 0.8676 |
MLLCNRKVPKTLNTCFILHIFTLLTLGVLVSGMPSKMVSFASQETLQRINNLLRGSANRD VDIIAEYLKKDDDDDGGDKDHHNIDIDPLPRRPSLTPDRQLLKVGLHGAISSDLEVCSNL TINEVLLKFPGSNAADAAVTQALCKGMVNFFNSGIGGGGYVVFSGKDDEDHLSIDFREKA PMDSHKFMFENCSLCSKIGGLAVGVPGELMGLYRLFKERGSGQVDWRDLIEPVAKLGSVG WQIGEALGATLELYEDVFLTLKEDWSFVLNSTHDGVLKEGDWIKRPALSNMLMELAKNGS VAPFYDPDHWIAKSMIDTVAKYNGIMNLQDVSSYDVHVTKPLSMKIRKGANFIPDNDMTV LTSSGSSSGAALLAALRIMDNFQNQEGGDYEKETTYHLLESMKWMASARSRLGDFEGEAL PKHIEEVLDPEWALKAVKSIKRNSQDGNFKTLENWTLYDPAYDINNPHGTAHFSIVDSHG NAVSLTTTINLLFGSLVHDPKTGVIFNNEMDDFAQFNKSNSFELAPSIYNFPEPGKRPLS STAPTIVLSELGIPDLVVGASGGSRITTSVLQTIVRTYWYNMPILETIAYPRIHHQLLPD RIELESFPMIGKAVLSTLKEMGYTMKEVFPKSVVNAIRNVRGEWHAVSDYWRKRGISSVY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YLR299W | 444 S | IKRNSQDGN | 0.994 | unsp | YLR299W | 444 S | IKRNSQDGN | 0.994 | unsp | YLR299W | 444 S | IKRNSQDGN | 0.994 | unsp | YLR299W | 540 S | KRPLSSTAP | 0.992 | unsp | YLR299W | 94 S | PRRPSLTPD | 0.992 | unsp | YLR299W | 164 S | YVVFSGKDD | 0.998 | unsp |