

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YMR274C | SP | 0.493080 | 0.505081 | 0.001839 | CS pos: 23-24. LYA-TS. Pr: 0.5523 |
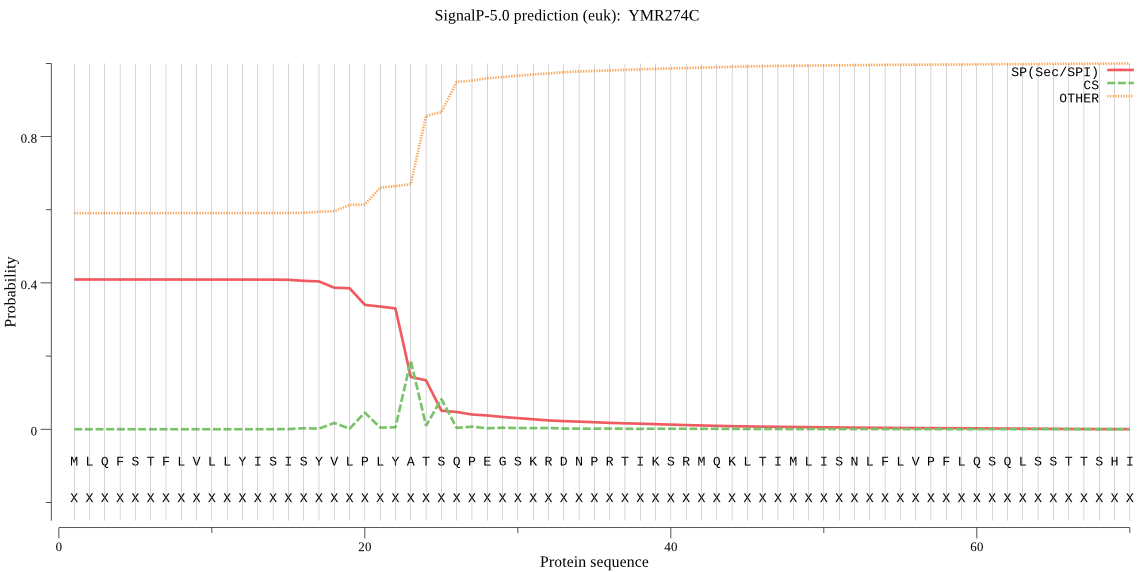
MLQFSTFLVLLYISISYVLPLYATSQPEGSKRDNPRTIKSRMQKLTIMLISNLFLVPFLQ SQLSSTTSHISFKDAFLGLGIIPGYYAALPNPWQFSQFVKDLTKCVAMLLTLYCGPVLDF VLYHLLNPKSSILEDFYHEFLNIWSFRNFIFAPITEEIFYTSMLLTTYLNLIPHSQLSYQ QLFWQPSLFFGLAHAHHAYEQLQEGSMTTVSILLTTCFQILYTTLFGGLTKFVFVRTGGN LWCCIILHALCNIMGFPGPSRLNLHFTVVDKKAGRISKLVSIWNKCYFALLVLGLISLKD TLQTLVGTPGYRITL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| YMR274C | 30 S | QPEGSKRDN | 0.997 | unsp | YMR274C | 71 S | TSHISFKDA | 0.998 | unsp |