

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
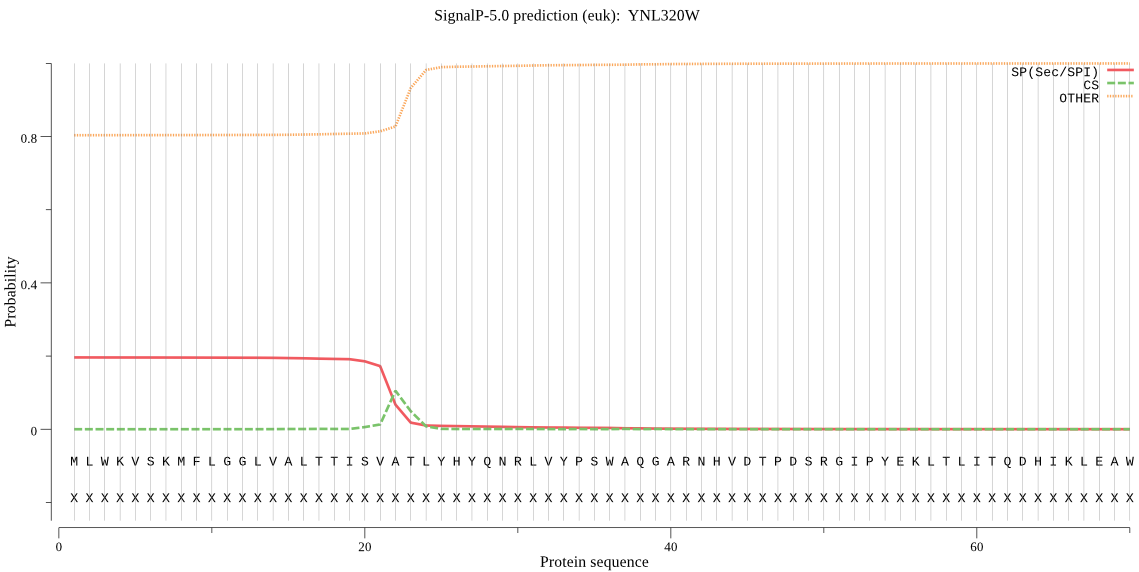
| YNL320W | SP | 0.327405 | 0.453301 | 0.219294 | CS pos: 22-23. SVA-TL. Pr: 0.5046 |
MLWKVSKMFLGGLVALTTISVATLYHYQNRLVYPSWAQGARNHVDTPDSRGIPYEKLTLI TQDHIKLEAWDIKNENSTSTVLILCPNAGNIGYFILIIDIFYRQFGMSVFIYSYRGYGNS EGSPSEKGLKLDADCVISHLSTDSFHSKRKLVLYGRSLGGANALYIASKFRDLCDGVILE NTFLSIRKVIPYIFPLLKRFTLLCHEIWNSEGLMGSCSSETPFLFLSGLKDEIVPPFHMR KLYETCPSSNKKIFEFPLGSHNDTIIQDGYWDIIRDFLIEKGFI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YNL320W | 147 S | DSFHSKRKL | 0.991 | unsp | YNL320W | 147 S | DSFHSKRKL | 0.991 | unsp | YNL320W | 147 S | DSFHSKRKL | 0.991 | unsp | YNL320W | 123 S | NSEGSPSEK | 0.995 | unsp | YNL320W | 125 S | EGSPSEKGL | 0.995 | unsp |