

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YOR003W | SP | 0.002910 | 0.997067 | 0.000023 | CS pos: 17-18. CLC-MI. Pr: 0.7718 |
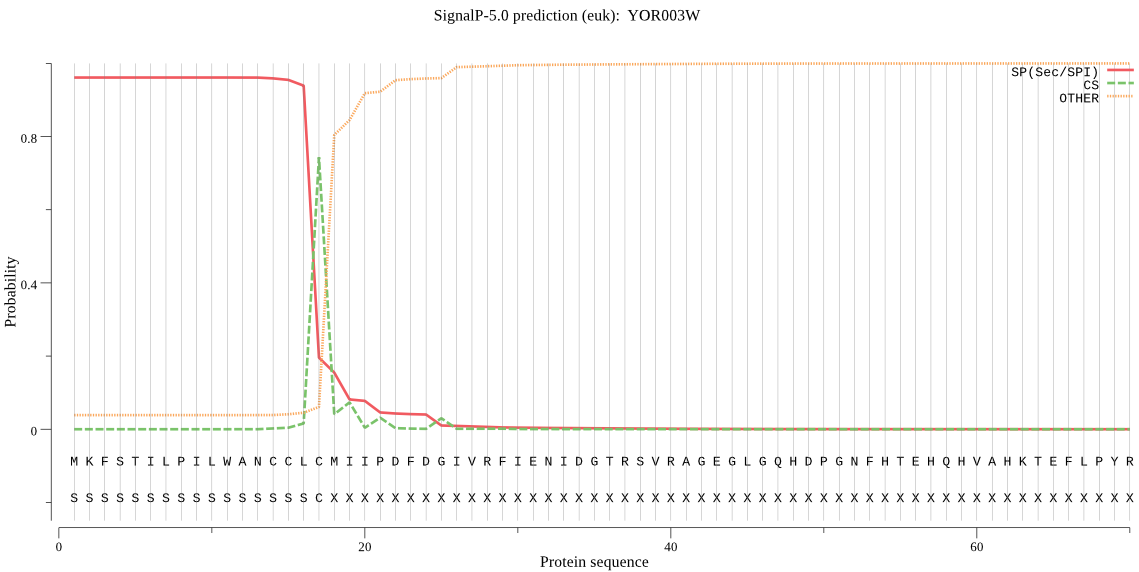
MKFSTILPILWANCCLCMIIPDFDGIVRFIENIDGTRSVRAGEGLGQHDPGNFHTEHQHV AHKTEFLPYRYVIVFNEDISLQQIQSHMQVVQKDHSTSVGKLTENDAFWRVISSSVSSKS QFGGIDNFFDINGLFRGYTGYFTDEIIKIISQDPIIKFVEQETTVKISNSSLQEEAPWGL HRVSHREKPKYGQDLEYLYEDAAGKGVTSYVLDTGIDTEHEDFEGRAEWGAVIPANDEAS DLNGHGTHCAGIIGSKHFGVAKNTKIVAVKVLRSNGEGTVSDVIKGIEYVTKEHIESSKK KNKEFKGSTANLSLGSSKSLAMEMAVNAAVDSGVHFAIAAGNEDEDACLSSPAGAEKSIT VGASTFSDDRAFFSNWGTCVDVFAPGINIMSTYIGSRNATLSLSGTSMASPHVAGILSYF LSLQPAPDSEFFNDAPSPQELKEKVLKFSTQGVLGDIGDDTPNKLIYNGGGKKLDGFW
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YOR003W | 184 S | LHRVSHREK | 0.998 | unsp | YOR003W | 184 S | LHRVSHREK | 0.998 | unsp | YOR003W | 184 S | LHRVSHREK | 0.998 | unsp | YOR003W | 437 S | NDAPSPQEL | 0.996 | unsp | YOR003W | 38 S | DGTRSVRAG | 0.991 | unsp | YOR003W | 117 S | SSSVSSKSQ | 0.997 | unsp |